Journal of Creation 20(3):66–70, December 2006
Browse our latest digital issue Subscribe
Lipid rafts: evidence of biosyntax and biopragmatics
Information theorist Werner Gitt has proposed that information is best understood within a multidimensional framework. This framework has five dimensions: the statistical, syntactic, semantic, pragmatic and apobetic. Current evolutionary views of information and biology are largely restricted to the statistical dimension. Alex Williams has recently argued that the inheritance of biological information should be understood within this multidimensional Gittian framework and has emphasised that extra-nuclear cell structure may contribute to the information content of cells. In this article I show that the emerging concept of ‘lipid rafts’ in cell membranes represents both a syntactic and a pragmatic information structure within the cell. As such they illustrate the existence of a higher order of organisation within the cell than traditionally understood and provide strong support for the Gittian creationist concept of how information is structured and inherited in biology.
* Terms marked with an asterisk are defined in the Glossary at the end of this article.

Werner Gitt has described information in terms of statistics, syntax, semantics, pragmatics and apobetics.1 Within this theory, the lower levels of information are hierarchically arranged in order to express the higher levels: i.e. statistics, syntax, semantics and pragmatics are arranged in such a way as to express the apobetics or purpose, within the organism (endowed by its Creator). In an analogy with linguistics, this is equivalent to words, sentences, paragraphs and context being arranged in such a way that the intended meaning (endowed by the author) may be conveyed, understood and acted upon.
This contrasts sharply with the evolutionary position, which denies that any purpose exists in biology. In the ‘selfish gene’ theory, for example, the ‘appearance of purpose’ is considered to be nothing more than a consequence of natural selection. Genes are believed to be the descendants of a hypothetical primordial ‘replicator’ that originated life and produced all of life’s variety simply by natural selection favouring different combinations in different environments. Information in this theory is nothing more than a serendipitous accumulation of statistical errors that happen to have survival value for the genes.
Conversely, the Gittian information framework, and creationists in general, interpret genetics and other biological processes in terms of their roles in enabling purpose to be fulfilled. The most important purposes expressed in biology are for organisms to reproduce after their kind, for humans to bear God’s image and worship Him, and to fill the earth and subdue it.
If Gitt’s information framework is applicable to biology then there should be evidence of information operating at the levels of syntax, semantics and pragmatics within biological systems. Alex Williams has recently described these features within a key area of genetics.2 He proposes that the nucleotide sequence found in DNA represents statistical information, the amino acids specified by codons represent a semantic arrangement, the sequence of amino acids represents a syntactic arrangement, and the function of gene products represents a pragmatic arrangement. This description is appropriate but is incomplete because gene products do not simply operate pragmatically but also form syntactic, semantic and pragmatic arrangements within ever more complex processes that make up the whole living organism.
In addition, Williams also highlighted the importance of structural modes of inheritance, including cellular architecture, as providing a basis for the stasis that is a major characteristic of inheritance and which forms part of the apobetic purpose of reproducing after like kind.3
In this article, I will show that the emerging concept of lipid rafts and their function as cell-signalling platforms can be understood as both a syntactic and a pragmatic arrangement of proteins and lipids and that this is derived from an interaction between genetic and epigenetic factors*. As such, they constitute a ‘molecular context’ or ‘cellular paragraph’ within which meaning is conveyed. Within the so-called ‘semiotic triad’ of object, sign and interpreter,4,5 lipid rafts form part of the interpretive machinery of the cell that allows the meaning of the symbol to be defined and connected to an appropriate response. Understanding the interpretative machinery of the cell is an important step in beginning to describe the way in which non-sentient entities, such as cells, translate abstract information into action without conscious will. The interpretative machinery of the cell closely resembles Gitt’s description of operational information.1 It is therefore apparent that lipid rafts form part of an operational information structure within the cell.
Lipid rafts
The term ‘lipid raft’ is used to describe areas of cell membranes that are selectively enriched in cholesterol, sphingolipids* and signalling proteins* . Since Simons and Ikonen brought attention to the possibility of their existence in 19976 there has been an explosion in efforts to detect and characterise these curious structures.7 They are believed to form functional domains on the outer leaflet of cell membranes that selectively include or exclude particular proteins depending on their levels of phosphorylation* , activation or the presence or absence of important ‘lipid tails’. In addition, lipid rafts are believed to somehow connect to intracellular signalling and structural proteins and influence their activities. Lipid rafts may thus concentrate members of different signalling cascades* within a discrete area of the cell allowing the components of the pathway to be in the right place at the right time.7
Do lipid rafts exist?
Although there are several lines of evidence that argue for the existence of lateral arrangements of cholesterol, sphingolipids and proteins,8 some of the techniques used to study them may artificially congregate these elements (e.g. detergent extraction)9,10 while other techniques have produced mixed results (e.g. fluorescent microscopy).11–14 Nevertheless, the weight of evidence seems to suggest that lateral organisation of the outer leaflet of the cell membrane occurs in vivo.
What are lipid rafts comprised of?
The plasma membrane* is composed of two phospholipid layers with the hydrophobic hydrocarbon* chain ‘tails’ facing each other and the hydrophilic* polar* ‘head groups’ facing the internal and external surfaces. Membranes have long been known to also contain proteins, cholesterol, sphingolipids (including the class of glycosphingolipids known as gangliosides), and other components. It has only recently been proposed that these various components do not exist completely randomly or evenly across the plasma membrane surface but that there is a level of lateral organisation. Thus there is evidence that sphingolipids and cholesterol are relatively concentrated in ‘rafts’ in the outer leaflet of the membrane and that proteins may have a preference for either the raft or non-raft regions depending on the biophysical properties of the protein in question. See figure 1.
This lateral arrangement is controlled in part by the levels of cholesterol and glycolipids, including gangliosides. Gangliosides are glycosphingolipids with one or more sialic acid* residues and are often used as markers of lipid raft domains. Their biosynthesis and regulation is complex (indeed it constitutes another cellular process that displays syntax, semantics and pragmatics!) and it is worth noting that they are not directly encoded in the genome since they are not proteins. They require the cooperative action of several enzyme complexes to construct them, direct them to their appropriate location and to maintain their presence in the outer plasma leaflet.15–22 The hydrophobic ceramide* tails, the hydrophilic sugar residues and the high levels of cholesterol mean that lipid raft domains have distinct physicochemical properties compared with neighbouring non-raft membrane.
What do lipid rafts do?
Lipid rafts have been implicated in a plethora of cellular processes. The common mechanism thought to underlie their function in most cases is their ability to preferentially concentrate or exclude molecules of cell signalling cascades.7 This ability is a result of the different biophysical properties of raft and non-raft membrane and the changes in chemical properties of proteins upon activation, phosphorylation or other events. For example, immunoglobulin E (IgE) signalling forms part of the allergic immune response and is activated when IgE binds to Fc receptors* (FcR) present on the plasma membrane of mast cells* . This probably increases the partitioning of the FcR into lipid rafts with subsequent cross-linking of FcR-bound IgE leading to phosphorylation of the FcR. This can then initiate secondary downstream signalling events including the release of histamine* .23 Other signalling pathways are proposed to operate in a similar manner with lipid rafts acting as concentrating platforms.24–26
Lipid rafts as biosyntax
Within written language systems, syntax refers to the set of rules that govern the way in which semantic objects (words) are positioned with respect to one another, how they may be joined together and how they may be modified while still retaining the intended meaning. For example the statement: ‘Does John like Rosalyn?’ differs from the statement ‘John does like Rosalyn!’ only in word order and punctuation. However, the change in order of the first two words changes the meaning (semantics) and purpose (apobetics) of the statement from being a question designed to elicit an answer to being a statement that answers a question. There is no statistical difference between the information content of the two statements (since they contain the same letters, words and spaces) but they differ at the level of syntax and consequently at the semantic, pragmatic and apobetic levels.

In a similar manner, lipid rafts can be seen to contribute to a molecular syntax within the cell. For example, it has been shown that in cultured cortical neurons* , the activation of the transmembrane signalling molecule ErbB4 by its ligand neuregulin (NRG) induces the translocation of ErbB4 and adaptor signalling molecules* into lipid raft domains from non-raft domains.27 Furthermore, this translocation into lipid rafts is required for NRG to exert its downstream effects, including activation of the protein kinase ERK. When lipid rafts are disrupted, the effects of NRG-induced activation of ErbB4 are not seen.
This situation is analogous to the sentences discussed above where the same words mean something different when the syntax is altered. The two conditions, ‘ErbB4 in lipid rafts’ and ‘ErbB4 not in lipid rafts’, differ only in the relative position of the relevant components of the system and not in the presence or absence of these components. The lipid rafts do not change the statistics of the information in the signalling pathway. Instead, they form a context in which the arrangements of the members take on defined meanings. Thus in these cultured cortical neurons, lipid rafts provide the molecular syntax in which NRG-induced activation of ErbB4 can bring about particular effects that do not occur in their absence (figure 2).
Lipid rafts as biopragmatics
Another example involving ERK signalling illustrates the way in which lipid rafts can act in the role of molecular pragmatics. In linguistics, semantics deals with the range of possible meanings of words and in biology it refers to the range of possible functions of molecules. Syntax, as we have seen above, deals with the different ways that components can be arranged to activate those meanings. Pragmatics, on the other hand, is all about context. An author identifies a particular intended meaning by the context in which he/she uses a word, and in biology, the particular function of a molecule is also specified by the context. In this example, the protein molecule CD45 has two quite different cellular functions depending on its arrangement within or outside of lipid rafts.
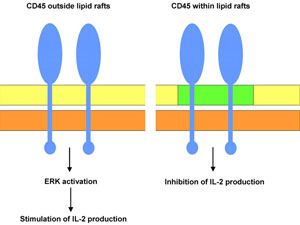
Zhang and colleagues have shown that membrane compartmentalisation of the transmembrane protein tyrosine phosphatase CD45 is an important feature in the regulation of T cell activation* in the mouse.28 These authors found that a proportion of CD45 is found within lipid rafts and a proportion is found outside lipid raft domains. The raft-localised CD45 was found to antagonise IL-2 production (IL-2 is a cytokine involved in several important immunological processes) while the CD45 located outside lipid rafts played a role in promoting IL-2 production via ERK activation.
There is evidence that during the stimulation of T cells, raft-associated CD45 is excluded from these domains. This presumably leads to less inhibition of IL-2 and greater stimulation of IL-2 via ERK. Thus a single molecule can have opposite effects (stimulate or inhibit IL-2 production) depending on the molecular context within which it operates (i.e. within or outside of lipid rafts) (figure 3).
The mechanisms linking CD45 signalling to IL-2 stimulation or inhibition are not well understood, but it is likely that the raft and non-raft membrane domains contain different proteins comprising distinct intracellular signalling cascades. Lipid rafts act as a component of this molecular context and so contribute to the pragmatics of the cell.
Inheritance of lipid rafts
Lipid rafts are components of cell membranes comprising of lipids and proteins. As such, they are passed on from parent cell to daughter cell during cell division as part of the cellular architecture that each cell inherits. In addition, the proteins that contribute to their formation and maintenance (e.g. the enzymes involved in ganglioside biosynthesis) are inherited both epigenetically as proteins in the lipid bilayer, and genetically as genes on chromosomes in the nucleus. This means that lipid rafts are passed on fully established and operational in an epigenetic manner, while the relevant information for their maintenance is passed on in a genetic manner. This illustrates how inheritance of the organised membrane structure forms part of the cellular basis of stasis of the created kind, as Williams has hypothesised.3
Cell autonomy
Syntax, semantics and pragmatics are used in language by intelligent beings according to conventions that allow purpose or apobetics, to operate. This requires an ongoing conscious tie between the language symbols and their referents. But the control and communication that operates within and between cells goes on autonomously without an ongoing conscious tie between symbols (e.g. DNA sequence, cell-signalling proteins, etc) and their referents (e.g. protein sequence, effectors of cell-signalling pathways, etc). Instead, the ties between object, sign and interpreter are established and maintained by an inherited information-based control system that operates according to biochemical laws.
Could such information-based cellular life have arisen via the ‘selfish gene’ replicator mechanism? It would seem not, because a gene can only use its information if it is part of a mechanism that contains transcription, translation and implementation facilities (i.e. a cell). So when lipid rafts are observed to function as syntactic and pragmatic information structures in cells it points to an intelligent rather than random original source.
Conclusion
The existence and operation of the lipid rafts in cell membranes can be seen as an implementation of syntax and pragmatics in order to enable apobetics to operate in the biological world in the absence of an ongoing conscious tie between object, sign and interpreter by the cell. It therefore supports the notion that the multidimensional framework of information of Werner Gitt is applicable to biological systems. Since this understanding of information requires an intelligent source, observing it in biological systems suggests their origin in an intelligent source rather than random, unintelligent statistical processes.
The Gittian view of information emphasises apobetics as the ultimate controlling factor in information-processing systems. This highlights the need to understand biology from the perspective of ‘apobetics-down’ (i.e. the purpose-orientated creationist perspective), rather than from the perspective of ‘statistics-up’ (i.e. the gene-orientated evolutionary perspective). There is a fresh need for a biblically derived apobetic theology of biology which provides a purpose-oriented framework for understanding biological processes and the nature of bioinformation. Such a theology would need to define the key purposes of creatures and relate these to biological processes, and would provide a philosophical foundation for studying the information present in organisms.
Glossary |
|
| Adaptor signalling molecules: | Proteins and other molecules that connect extracellular signals to intracellular signalling cascades. |
| Ceramide: | an N-acyl sphingosine which forms the lipid portion of glycosphingolipids. |
| Cortical neurons: | nerve cells from the cortical region of the brain. |
| Epigenetically: | information inherited via genes is inherited genetically. Information inherited in any other way is inherited epigenetically. |
| Fc receptors: | the portion of an immunoglobulin molecule that initiates its effector functions (e.g. complement activation, opsinisation, etc). |
| Histamine: | Derived from an amino acid, histamine is a chemical messenger involved in initiating many of the events involved in acute inflammation. |
| Hydrocarbon: | an organic molecule which consists of carbon and hydrogen atoms. |
| Hydrophilic: | having an affinity for water. By contrast, hydrophobic means lacking an affinity for water. |
| Mast cells: | an immune cell residing in connective tissue. It releases several substances, including histamine, involved in acute inflammatory reactions. |
| Phosphorylation: | the addition of a phosphate group to a protein molecule. This often alters protein structure and function and acts as a regulatory switch. |
| Plasma membrane: | the lipid layer which comprises the outer wall of each individual cell. |
| Polar: | the hydrogen bonding end of the phospholipid molecule, which is hydrophilic. |
| Sialic acid: | A 9 carbon acidic sugar. It is present on many glycolipids and glycoproteins and is responsible for much of the negative charge on cell surfaces. |
| Signalling cascades: | series of reactions which occur as a result of a single stimulus. |
| Signalling proteins: | proteins which contribute to the transfer of information from cell to cell. |
| Sphingolipids: | structural lipids derived from sphingosine (a long chain amino alcohol). |
| T cell activation: | under appropriate conditions, stimulation of inactive immune T-lymphocytes results in them becoming activated and capable of assisting in the immune response. |
References
- Gitt, W., In the Beginning was Information Christliche Literatur-Verbreitung, Bielefeld, Germany, 1997. Return to text.
- Williams, A., Inheritance of biological information—part I: the nature of inheritance and of information, Journal of Creation 19(2):29–35, 2005. Return to text.
- Williams, A., Inheritance of biological information—part III: control of information transfer and change, Journal of Creation 19(3):21–28, 2005. Return to text.
- Barbieri, M., The Organic Codes: The Birth of Semantic Biology, peQuod, Ancona, Italy, 2005. Return to text.
- Williams, A., Inheritance of biological information—part II: redefining the ‘information challenge’, Journal of Creation 19(2):36–41, 2005. Return to text.
- Simons, K. and Ikonen, E., Functional rafts in cell membranes, Nature 387:569–572, 1997. Return to text.
- Simons, K. and Toomre, D., Lipid rafts and signal transduction, Nat. Rev. Mol. Cell Biol. 1:31–41, 2000. Return to text.
- Chamberlain, L.H., Detergents as tools for the purification and classification of lipid rafts, FEBS Lett. 559:1–5, 2004. Return to text.
- Heerklotz, H., Triton promotes domain formation in lipid raft mixtures, Biophys. J. 83:2693–2701, 2002. Return to text.
- le Maire, M., Champeil, P. and Moller, J.V., Interaction of membrane proteins and lipids with solubilizing detergents, Biochim. Biophys. Acta 1508:86–111, 2000. Return to text.
- Kenworthy, A.K., Petranova, N. and Edidin, M., High-resolution FRET microscopy of cholera toxin B-subunit and GPI-anchored proteins in cell plasma membranes, Mol. Biol. Cell 11:1645–655, 2000. Return to text.
- Mayor, S., Rothberg, K.G. and Maxfield, F.R., Sequestration of GPI-anchored proteins in caveolae triggered by cross-linking, Science 264:1948–1951, 1994. Return to text.
- Pralle, A., Keller, P., Florin, E.L., Simons, K. and Horber, J.K., Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells, J. Cell Biol. 148:997–1008, 2000. Return to text.
- Zacharias, D.A., Violin, J.D., Newton, A.C. and Tsien, R.Y., Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells, Science 296:913–916, 2002. Return to text.
- Bieberich, E. and Yu, R.K., Multi-enzyme kinetic analysis of glycolipid biosynthesis, Biochim. Biophys. Acta 1432:113–124, 1999. Return to text.
- Bieberich, E., Tencomnao, T., Kapitonov, D. and Yu, R.K., Effect of N-glycosylation on turnover and subcellular distribution of N-acetylgalactosaminyltransferase I and sialyltransferase II in neuroblastoma cells, J. Neurochem. 74:2364, 2000. Return to text.
- Giraudo, C.G., Daniotti, J.L. and Maccioni, H.J., Physical and functional association of glycolipid N-acetyl-galactosaminyl and galactosyl transferases in the Golgi apparatus, Proc. Natl. Acad. Sci. U.S.A 98:1625–1630, 2001. Return to text.
- Giraudo, C.G. and Maccioni, H.J., Ganglioside glycosyltransferases organise in distinct multienzyme complexes in CHO-K1 cells, J. Biol. Chem. 278:40262–40271, 2003. Return to text.
- Gu, X., Preub, U., Gu, T. and Yu, R.K., Regulation of sialyltransferase activities by phosphorylation and dephosphorylation, J. Neurochem. 64:2295–2302, 1995. Return to text.
- Martina, J.A., Daniotti, J.L. and Maccioni, H.J., GM1 synthase depends on N-glycosylation for enzyme activity and trafficking to the Golgi complex, Neurochem. Res. 25:731, 2000. Return to text.
- Pohlentz, G., Klein, D., Schwarzmann, G., Schmitz, D. and Sandhoff, K., Both GA2, GM2 and GD2 synthases and GM1b, GD1a and GT1b synthases are single enzymes in Golgi vesicles from rat liver, Proc. Natl. Acad. Sci. U.S.A 85:7044–7048, 1988. Return to text.
- Tettamanti, G., Ganglioside/glycosphingolipid turnover: New concepts, Glycoconj. J. 20:301–317, 2004. Return to text.
- Goitsuka, R. et al.,A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation, Int. Immunol. 12:573–580, 2000. Return to text.
- Cheng, P.C., Dykstra, M.L., Mitchell, R.N. and Pierce, S.K., A role for lipid rafts in B cell antigen receptor signaling and antigen targeting, J. Exp. Med. 190:1549–1560, 1999. Return to text.
- Janes, P.W., Ley, S.C., Magee, A.I. and Kabouridis, P.S., The role of lipid rafts in T cell antigen receptor (TCR) signalling, Semin. Immunol. 12:23–34, 2000. Return to text.
- Wary, K.K., Mariotti, A., Zurzolo, C. and Giancotti, F.G., A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth, Cell 94:625–634, 1998. Return to text.
- Ma, L. et al., Ligand-dependent recruitment of the ErbB4 signalling complex into neuronal lipid rafts, J. Neurosci. 23:3164–3175, 2003. Return to text.
- Zhang, M. et al.,CD45 signals outside of lipid rafts to promote ERK activation, synaptic raft clustering, and IL-2 production, J. Immunol. 174:1479–1490, 2005.



Readers’ comments
Comments are automatically closed 14 days after publication.