Journal of Creation 25(3):23–26, December 2011
Browse our latest digital issue Subscribe
The junk DNA myth takes a well-deserved hit
A review of The Myth of Junk DNA by Jonathan Wells
Discovery Institute Press Seattle, WA, 2011

The history and status of the myth
One of the greatest evolutionary frauds of recent times is the myth that eukaryotic genomes, particularly the human genome, are largely composed of meaningless ‘junk’ DNA sequences that serve no biological purpose. While many of the actual researchers working in the vast field of genomics now realize that virtually the entire genome is functional,1 a handful of influential and popular bio-science authors still authoritatively proclaim the fraudulent myth of ‘junk DNA’. Thus, the errant junk DNA myth is still widely perpetuated even though several decades of hard-hitting research profoundly declares otherwise.
The problem is that the general public doesn’t often read the journal-based scientific literature, but instead rely on popular science authors to distill the pertinent research data into a more easily understood form. In this process, the truth about non-coding DNA is withheld and distorted by the popular media technical outlets. Unfortunately, the scientific technocracy of our time uses the junk DNA myth as a key component to perpetuate the overall myth of Darwinian evolution. A new book by molecular biologist and author Dr Jonathan Wells utterly and thoroughly destroys the cherished junk DNA paradigm. It is appropriately titled The Myth of Junk DNA.
The book begins with a brief and concise background exposé on the scientific failure of Darwinian evolution. This is done by skillfully quoting a variety of key evolutionists who indicate that, ever since the days of Darwin, no clear evidence has been discovered to support Darwinian evolution. Also mentioned is the failure of the evolutionary tree-of-life model in light of overwhelming DNA evidence emerging from the many recent genome projects. The concept of ubiquitous intelligent design found in living systems as counter-evidence to Darwinian evolution is also briefly summarized. Most importantly, Wells shows that the Darwinian paradigm is historically propped up by a handful of fraudulent scientific models called “icons of evolution”. In the past, these icons have been represented by various discredited paradigms such as Ernst Haeckel’s fraudulent embryology models. In more recent times, one of the key icons of evolution is the (false) concept of junk DNA. For those interested in exploring this concept of evolutionary icons in more detail, Wells has written a book appropriately titled Icons of Evolution.2
After showing how fraudulent icons have historically been used to prop up the Darwinian paradigm, Wells then illustrates the overall nature of the junk DNA issue before addressing in detail the actual genetic data. In doing this, he analyzes the widely distributed rhetoric of the leading academic technocrats and front men who foster the junk DNA myth. These evolutionary luminaries are Richard Dawkins, Michael Shermer, Ford Doolittle, Carmen Sapienza, Leslie Orgel, Francis Crick, Kenneth Miller, Douglas Futuyma, Francis Collins, Philip Kitcher, Jerry Coyne and John Avise. Wells provides many complete quotes derived from popular books and scientific reviews that provide ample evidence that the junk DNA myth is still alive and well.
The general consensus from all of the junk DNA rhetoric is that much of the genome which does not directly code for protein represents evolutionary vestiges of viruses, defunct genes, and other repetitive sequences dragged along through evolutionary history. Such regions are postulated to be ‘neutral’, for the most part, having experienced minimal levels of negative selection over evolutionary time. One of the more common arguments coming from this camp is the idea that a creator God would never have filled genomes with such large amounts of seemingly useless DNA. Therefore, when the evolutionary technocratic elite wave their magic wand and proclaim most of the non-coding sections are ‘useless’, that somehow removes any vestige of intelligent design. The current problem for the technocracy, however, is that Wells’ book does an excellent job of bringing the scientific data out in the open to counter such nonsense.
The experimental data destroys the myth
With the exception of several summary and discussion chapters and some important appendix material, the bulk of The Myth of Junk DNA lays out in sufficient and understandable detail the key published data unequivocally supporting a full-function view of the genome. Wells provides five well-organized chapters that present this data in a logical progression. One noteworthy point is that for each line of research into some aspect of noncoding DNA function, Wells uses a historical progression showing how the original discoveries open up a certain field and are then followed up by multiple research projects over time. Interestingly, many of the initial discoveries that launched a certain research trajectory that eventually helped nullify some errant aspect of the junk DNA fraud have one thing in common; they occurred in the 1980s or 1990s—the same period of time in which the myth is first widely initiated.
Non-coding DNA in the genome is widely transcribed
After a brief genome primer and after establishing the fact that nearly all of the genome is transcribed (RNA copied from DNA segments), Wells shows that many segments of so-called junk DNA are very similar across a wide variety of creatures. According to evolutionary reasoning, these sections should be heavily mutated and variable since they are supposedly non-functional and not actively undergoing purifying selection. However, the high level of similarity in non-coding DNA segments between species, usually called “sequence conservation”, is actually a strong indicator of its functionality in an evolutionary sense. Darwinian evolution errantly predicts that these regions of the genome should be riddled with mutations because they are non-functional junk.
A major argument indicating functionality of so-called junk DNA is the fact that diverse classes of non-protein-coding DNA are actually required as templates to make many different types of non-protein-coding RNAs that are known to regulate virtually all levels of gene activity in the genome. These RNAs are broadly classified as regulatory RNA with many different types of sub-classes. Interestingly, many of these RNAs are transcribed from the reverse (antisense) strand of the double-stranded DNA helix to produce RNAs that will base-pair in a complementary fashion to protein-coding RNAs transcribed from the forward (sense) strand. This can provide rapid regulation in several ways; to either keep messenger RNAs (mRNAs) from being degraded (preserving their presence in the nucleus) or keeping them from being translated (protein production) in order to down-regulate expression levels.
During the course of explaining these concepts, the publicly-funded ENCODE project (Encyclopedia of DNA Elements) is mentioned. ENCODE was a critical public research effort that discovered how much of the genome, even the parts that do not directly code for protein, is regulatory in some way. In fact, the regulation of gene expression by the wide variety of RNAs produced from non–protein-coding DNA sequence are now being perceived as more important targets of study than the actual protein-coding sections of the genome. The wide diversity of noncoding RNAs (ncRNAs) in a cell affects virtually all aspects of growth, development and physiology. The protein-coding sections of the human genome, comprising less than 3% of the whole, are somewhat analogous to the raw materials (bricks, boards, wire, etc.) one would use in a building construction project. It is the intelligent oversight, implementation and usage of these raw materials that makes the building take shape and function, and that seems to be what ncRNAs are used for.
Splicing code
After showing how the genome should be viewed as globally functional and not a sea of junk, Wells then gets into one of the most amazing features of the genome—the splicing code. This paradigm has been progressively defined over the past several decades, but has recently come together in more detail. Researchers claim that the number of human protein-coding genes is between 20,500 and 25,000,3 yet, at present, the NCBI database contains about 2.5 million human protein sequences. When accounting for entries that likely represent the same protein (based on amino acid sequence similarity), about 1.4 million different human proteins have been characterized to date. The paradox is that the number of proteins discovered greatly outnumbers the current estimate for the total number of protein-coding genes found in the human genome. Obviously, something very complicated is going on in a regulatory sense, roughly akin to the way a computer programmer practices code optimization and re-use. However, what has been discovered through the splicing code is much more complicated than any computer system developed by man.
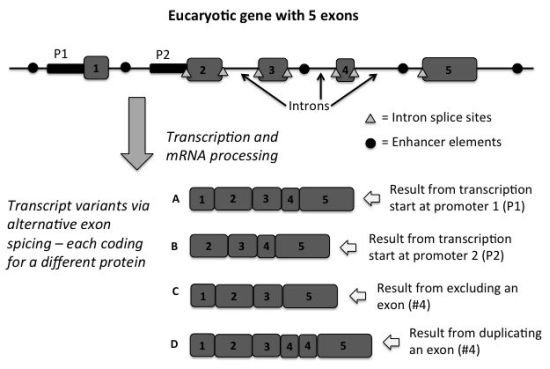
Eukaryote genes are interrupted by non-coding areas that are spliced out after RNA transcripts of the gene are produced. These are called introns. The coding regions are called exons. Scientists discovered early on in the genomics era that some genes exhibit alternative splicing, where exons are either omitted or added in the linear sequence of a transcript, allowing for the production of different proteins from the same sequence (figure 1). The great disparity between the number of protein-coding genes and the number of proteins produced appears to be controlled to a large extent by the splicing code.
As Wells clearly shows the reader, so-called junk DNA plays a comprehensive and immense regulatory role in this process. Not only does non-coding DNA contribute to exon selection and placement decisions, but it also controls the recognition and activity of alternative transcriptional start sites, splice site recognition, transcriptional/translational cues, transcript processing/transport, genome architecture, nuclear membrane architecture, etc. Thus, a large list of protein, DNA, RNA, and cell physiology factors combine in various configurations to produce different transcript and protein production activity. If there ever was a fool-proof example from the world of biology to unequivocally prove intelligent design, this is it. The Myth of Junk DNA does a good job of acknowledging and explaining most of these issues and Wells always provides the appropriate background information for each concept so that it is well understood.
Pseudogenes
Another critical topic related to the whole junk-DNA myth is the widely misunderstood subject of pseudogenes. The false idea that pseudogene regions of the genome represent cryptic, dysfunctional genes has long been a prime tool of the evolutionary fraudsters. The book does a fantastic job of first bringing the reader up to speed on the different classes of pseudogenes and then showing how popular science authors have presented their errant and misleading concepts to the public. After laying this groundwork, the historical scientific research is chronologically and quite thoroughly addressed. The infamous controversy surrounding the topic of the vitamin C human pseudogene is even addressed, but in the appendix in its very own chapter, which was an appropriate way to handle the issue and preserve the flow of the main text. As accurately illustrated, pseudogene sequence in many regions of the genome has been proven to be actively transcribed via both the minus and plus strands. These types of transcripts are then able to play a diversity of roles in both the up regulation and down regulation of a wide variety of genes that they are homologous at some level.
One possibility that is particularly intriguing, yet not well-covered in the book, is the putative activation of pseudogene sequence via genomic splicing, wherein regulatory genomic sequence would be literally moved or copied and then spliced into position near the pseudogene. The genome is not as static, in a linear structural sense, as scientists had previously imagined. Recent research has shown that during embryo development, a wide variety of genomic rearrangement takes place, presumably to facilitate a variety of dynamic transcriptional activity.4 In fact, Wells does show how information in the genome occurs in at least three different levels (see below).
Transposable elements
One class of DNA that has also been the target of evolutionary propagandists is the diverse group of DNA features called transposable elements (TEs), sometimes referred to as “jumping genes”. Many scientists have stated that all of the subclasses in this diverse group were merely genomic baggage conferred by ancient ancestral viruses that infested our DNA and have served no other purpose than to bloat and expand the genome with meaningless DNA fragments. However, the past several decades of research now show that every identified class of TEs (LINEs, SINEs, ERVs and DNA-transposons) all have important roles in the function of the genome during development, growth, and day-to-day physiological activity. Wells discusses the history of these discoveries along with detailed information about each sub-class of TE and its currently known functional characteristics. Interestingly, all of these TEs are now known to have multiple functions depending on the type of cell and its activity. Rather than cryptic viral genome contaminants, it is now clear that TEs are absolutely critical to genome function and survival. TEs apparently also play important roles in cell stress responses and other protective measures. The Myth of Junk DNA clearly shows how the various classes of TEs present in the genome are not useless trash, but indispensible functional factors.
Non-coding structural genomic information
Besides the linear-based DNA sequence information, Wells illustrates an important concept that is often not discussed in scientific circles because of its overtly design-rich overtones. There are three levels of non-sequence-based information inherent in DNA structure:
- DNA is not randomly strewn about the nucleus, but instead is packaged into various organizational levels. In addition, chemical modifications are added to the DNA molecule itself. These include nucleotide methylation and histone acetylation. Physical factors such as the stresses caused by torsion add to the complex structure of DNA in vivo. The levels of packaging and chemical modification greatly affect gene function and access by a wide variety of regulatory molecules. These modifications are heavily influenced by the presence of a wide variety of non-coding DNA features.
- The 3-dimensional orientations assumed by DNA are often referred to as “local genome topology”. In any given local region of the genome, it may contain looping structures, structural RNA integration, and specific nuclear matrix attachments. These local topological features and configurations influence gene function/regulation and are facilitated and made possible by the non-coding DNA regions of the genome.
- The global configuration of the genome is another key subcellular feature. The dynamically configured overall 3-dimensional genome structure depends on the cell type and its associated stimuli and cues. In such a scenario, certain chromosomes and chromosomal regions are dynamically configured to occupy specific functional domains. These 3-dimensional features and configurations are also facilitated and made possible by the non-coding DNA regions of the genome.
Current status of junk DNA propaganda
While Wells properly addresses the history of the junk DNA myth and its associated rhetoric at the beginning of the book, he revisits the subject again at the end of the book, providing more detail on the current state of dogma. He shows how many of the popular science authors are spinning a myth largely disconnected from real research and relying on antiquated concepts. In fact, given the current state of knowledge, it is becoming increasingly apparent that these evolutionary evangelists are more concerned about spreading their dogma and disinformation than an accurate view of modern genetics.
Perhaps the most interesting of these cases mentioned is that of NIH Director Francis Collins, who headed the public sector of the Human Genome Project from 1993 to 2007. While Collins has made a number of errant dogma-driven junk DNA statements during this tenure, he has been forced in recent years to correct his position on the issue. Some of these corrections may largely be due to his co-authoring of key research papers that demonstrate function for non-coding DNA. Nevertheless, the organization that Collins founded to meld Christian faith with evolutionary dogma (Bio-Logos5) still perpetuates the junk DNA myth through several of its chief leaders and writers. As Wells thoroughly demonstrates, the leading junk DNA peddlers seem more interested in propagating their errant dogma than keeping the populace abreast of all the exciting discoveries that show how wonderfully designed and complex genomes really are.
The Myth of Junk DNA—a reference for creation biologists
Not only was this book a highly informative and fairly thorough read on the subject in question, but it is also a great resource for creationist and other scientists to keep on hand. Wells provides an excellent bibliography citing in full the key journal paper resources for each chapter. While key milestone research papers for each topic are referenced, many of the leading journal-based review papers on a particular topic are also listed. Review papers in journals are compilations of experimental discoveries in a particular field produced by leading researchers in that field. These are excellent resources for delving more deeply into each of the topics that Wells covers in his book. In other words, this book is an excellent resource to allow the reader to pursue any topic of interest covered in the book in deeper detail.
Conclusion
This book is a must-read for every creation biologist, and an invaluable resource to keep on hand. I would also highly recommend this book as a means to clearing up any misconceptions that people have developed in response to the errant dogma of junk DNA perpetuated by the various popular science authors.
Although I do not consider myself an expert on every field and subfield in the vast sea of research now occurring in the area of non-protein-coding regions of genomes, I believe Wells did a fantastic job of covering and mentioning virtually all of the key areas. One suggestion for future editions would be to add a few paragraphs on the transcription of telomeric repeats to produce molecules called TERRAs—telomere repeat RNAs.6 The field of TERRA research is exploding in a variety of directions. It has been recently reported that TERRAs play important roles in such diverse areas as chromatin remodeling, cellular differentiation, RNA surveillance machinery, X chromosome inactivation, centromere stability, and cellular immune responses.7,8
My final suggestion is that I would hope that Dr Wells considers this book an ongoing project. Given the immense amount of research in this field and the new and exciting developments which emerge virtually every week, the book should be regularly revised every couple of years to keep the issue at the forefront in the hotly contested fields of intelligent design and creation science. This is particularly important given the fraudulent rhetoric actively promulgated by theistic evolutionists and popular science authors.
References
- Williams, A., Astonishing DNA complexity demolishes neo-Darwinism, J. Creation 21(3):111–117, 2007. Return to text.
- Wells, J., Icons of Evolution, Regnery Publishing, Washington, DC, 2004; www.iconsofevolution.com/. Return to text.
- Clamp et al., 2007, Distinguishing protein-coding and noncoding genes in the human genome, Proc. Natl. Acad. Sci. 104:19428–19433; see also review: Truman, R., What biology textbooks never told you about evolution, J. Creation 15(2):17–24, 2001. Return to text.
- Probst, A.V. and Almouzni, G., Pericentric heterochromatin: dynamic organization during early development, Differentiation 76:15–23, 2007. Return to text.
- See creation.com/biologos for detailed critique and the apostasy to which theistic evolutionary compromise logically leads. Return to text.
- Tomkins, J. and Bergman, J., Telomeres: implications for aging and evidence for intelligent design, J. Creation, 25(1):86–97, 2011; creation.com/journal-of-creation-251. Return to text.
- Luke, B. and Lingner, J., TERRA: telomeric repeat-containing RNA, EMBO J. 28:2503–2510, 2009. Return to text.
- Farnung, B.O. et al., Promoting transcription of chromosome ends, Transcription 1:140–143, 2010. Return to text.



Readers’ comments
Comments are automatically closed 14 days after publication.