Journal of Creation 28(1):91–97, April 2014
Browse our latest digital issue Subscribe
Human genome decay and the origin of life

Recent reports on the human genome provide powerful support for the biblical history of Creation and Fall. We are unable to reproduce ourselves without making multiple genome copying errors every generation. As a result our genomes are decaying towards extinction from copy errors alone. However, they make up only 0.1% of the total mutation burden, so 99.9% of that burden must have come from other causes. When decay in copy fidelity is projected backwards in time it reaches perfection around 4,000 BC, and when projected forwards, extinction from copy errors alone occurs in thousands, not millions, of years. Naturalistic models of copy fidelity decay cannot explain the data because they rapidly collapse into ‘error catastrophe’ (mutation meltdown). Only an Intelligent Design (ID) model with perfect original copy fidelity, plus ongoing maintenance and repair, can persist for thousands of years to fit the data. The ID model produced no copy errors at all for the early human generations, few until the time of Abraham, and a total today of only ~4,000 compared with our total burden of ~3 million. Darwinian scenarios requiring an extremely remote origin of life with a low primordial copy fidelity are absolutely excluded by this data!
In the original creation, before the Fall of man, there was no death or suffering. The Word of God through whom the world was made (John 1:3) also upheld the world perfectly (Hebrews 1:3). We are given a glimpse of this condition in the story of Daniel’s friends in the fiery furnace (Daniel 3). Shadrach, Meshach and Abednego went into the fiery furnace, walked around in it, and came out again without even the smell of burning upon them. When a single human hair or a single animal hair (e.g. in their garments) is burned the protein breakdown products create a pungent smell. This description is telling us that not even a single hair on their bodies or clothing had been harmed.
From this general principle of perfect divine upholding we can infer that the human genome would have been copied with 100% accuracy into Adam and Eve’s descendants had their fall into sin not occurred. But the Bible tells us that God foresaw the Fall and made provision for it (e.g. Ephesians 1:3–14; 2 Thessalonians. 2:13; 2 Tim. 1:9; Revelation 13:8). This must have included sustaining the functionality of organisms and their offspring through many mutation-prone generations after the Fall. In this article I examine just one part of the process of human genome decay—the errors due to genome copying. I then extrapolate backwards and forwards in time and compare it with the Bible and with Darwinian scenarios of origin and destiny.
Human genome decay
In his landmark book, Genetic Entropy & the Mystery of the Genome, geneticist Dr John Sanford clearly demonstrates that human genomes are decaying at an unstoppable rate, a principle he calls ‘genetic entropy’.1 The reason for the decay is that natural selection can only remove the severely deleterious mutations from the gene pool. The vast majority of mutations are only mildly deleterious, or they have no detectable effect at all, so they are passed on from one generation to the next and accumulate continuously. Sanford includes a model calculation that predicts our species will become extinct in about 300 generations (6,000 years, with a generation time of 20 years). In an earlier article,2 I used some other lines of evidence to illustrate genome decay and I included two quantitative methods to estimate the timescale to extinction. The estimated times ranged from 1,000 to 1,500,000 years, with a possible average of 30,000 years.
A far more important development occurred with the release of Sanford and colleagues’ powerful computer simulation program Mendel’s Accountant.3 This allows greater refinement in predicting the fate of mutations in populations and the results agree with Sanford’s earlier work. There appear to be no (realistic) evolutionary models in the scientific literature that contradict these results. This overwhelmingly negative evidence clearly contradicts Darwinian expectations, but clearly and dramatically fits the biblical record of the Fall.
DNA copying
The Author of life’s solution to the genome degeneration problem was to provide cells with not just one but a multitude of DNA maintenance and repair mechanisms. Together they ensure a relatively healthy life in the current generation and the prospect of viable offspring for many generations to come.
If such mechanisms had not been present, life would have become extinct very quickly through the multiplication of errors—a condition called ‘error catastrophe’, which I shall illustrate shortly. The technical term for maintaining genome quality during reproduction is ‘DNA copying fidelity’ and the enzyme systems that do the copying are called ‘DNA polymerases’. Copy fidelity maintenance mechanisms include proofreading, numerous kinds of error correcting systems, and error accumulation checkpoints.
According to Thomas Kunkel, a specialist in this field:
“DNA copying fidelity is an important area of scientific research … because the balance between correct and incorrect DNA synthesis is relevant to a great deal of biology. High fidelity DNA synthesis is beneficial for maintaining genetic information over many generations and for avoiding mutations that can initiate and promote human diseases … . Low fidelity DNA synthesis is beneficial for the evolution of species, for generating diversity leading to increased survival of viruses and microbes when subjected to changing environments, and for the development of a normal immune system. What was not appreciated [by the pioneers in this field] … was the large number and amazing diversity of transactions involving DNA synthesis required to faithfully replicate genomes and to stably maintain them in the face of constant challenges from cellular metabolism and the external environment.”4
Copy fidelity varies with the different DNA copying systems used, with the different kinds of errors involved, and with the different stages in a cell’s life cycle. Error rates seem to vary across almost all possibilities, from 1 per nucleotide copied to about 1 in 10–100 million nucleotides, depending on the copy-repair system. Cells also appear to have the ability to combine several diverse copy-repair systems in different ways to achieve cooperatively a greater fidelity than any one system can achieve individually.5 Overall it appears to be impossible for our cells to copy the 3 billion nucleotides in our genome without error.
A recent study of autozygous DNA in whole genomes of five genealogically well-defined Hutterite parent-offspring trios yielded a single nucleotide mutation rate of 1.2 per hundred million base-pairs per generation.6 In a genome of 3 billion base-pairs that amounts to 36 single nucleotide changes (SNPs)7 per person per generation. This figure is smaller than previously measured rates (perhaps because of the investigator’s narrow focus) but will suffice for present purposes. Mutation rates must have varied considerably in the past because a recent study of protein coding genes showed that about 86% of deleterious SNPs have accumulated in the last 200 generations.8
Looking back to the origin of life
In the approximately 6,000 years since creation, humans have gone through roughly 250 generations. In each fertile female the embryonic sex cells undergo about 23 cell generations to produce approximately 7 million primary eggs in the developing ovary,9 but this number is then selected back to about 1–2 million in the mature ovaries.10 No more eggs are produced during the female’s lifetime. Males, on the other hand, continue to produce new sperm throughout life so they continue to accumulate mutations with age. In the Hutterite family study mentioned above, 85% of SNPs came from fathers and only 15% from mothers. Female DNA plays a foundational role in maintaining the viability of life in the long term because it is the egg-cell, with its high-fidelity maternal DNA, that becomes the first cell of the offspring.
Males only contribute chromosomes. When Adam named his wife Eve, “because she would become the mother of all the living” (Genesis 3:20), he spoke a biological truth that would only become known to science 6,000 years later. The cells of all our bodies are the lineal descendants of the cells of Eve’s body, but not of Adam’s. The importance of this point will be seen shortly.
Our current human population has therefore gone through about 23 × 250 = 5,750 germ-cell generations in the maternal line since creation. According to the findings of The 1000 Genomes Project we have each accumulated on average about 3.6 million SNPs in that time.11 Some of these would have been built into our original parents (Adam and Eve) to provide a pool of potentially useful variation for later generations to draw on. A recent example is the discovery that a single nucleotide change in ethnic Tibetans (compared with Han Chinese) has allowed them to cope with the chronically low oxygen levels that occur on the high Tibetan plateau.12 In the recent study of protein coding genes mentioned above, the 86% of deleterious SNPs that accumulated in the last 200 generations would amount to approximately 3 million if applied to this average genome figure (3.6 million x 0.86 = 3 million). Several ancient human genomes have been sequenced and the one with the fewest SNPs (i.e. our closest estimate of the built-in variation) at about 450,000 belonged to a Paleo-Eskimo.13 It seems reasonable therefore to assume that something like 3 million of our SNPs have accumulated since creation.14 With our 3-billion-nucleotide genomes carrying 3 million SNPs then one measure of the current state of our genomic health is that we each carry approximately 1 error per thousand nucleotides.
Genome degeneration modelling
When the principle of genetic entropy is extrapolated backwards into history we must expect to see higher quality genomes and higher fidelity of reproductive copying the further back we go in time. We could perhaps use Mendel’s Accountant to simulate this process but it is more instructive for the general reader to see the process at work in some simpler models.
The simplest calculation we could do is multiply the number of mutations accumulating today per generation (36 in the Hutterite study) by the number of generations since creation (~250) which gives us 9,000 mutations in total. This is only 0.3% of the 3 million SNPs measured in genome studies. The huge difference suggests we need a more complex model to represent what is going on.
Copy fidelity between any two generations is clearly a different thing to mutation accumulation across many generations, so we can represent them as two separate components in a model such as the following:
Genome decay = copy fidelity decline + mutation accumulation from other sources
When parents today have children, they pass on the millions of SNPs inherited from their ancestors plus 36 new ones (in the Hutterite case) that have accumulated during their own generation.15 Suppose, however, that one parent is exposed to excessive radiation. That parent will pass on an extra burden of mutations that has nothing to do with genome copy fidelity. Suppose, again, that another parent suffers damage to their genome copying mechanism so that its fidelity declines to 1 error per million nucleotides in just that one generation. That parent will pass on the millions of SNPs inherited from earlier generations, plus the extra copy errors—3 billion nucleotides multiplied by 1 error per million nucleotides = 3,000 new mutations from just that one generation, plus the likelihood of greater copy error rates in subsequent generations.
Copy fidelity multiplies errors between generations, while mutations from other sources (radiation, diet, smoking, environment, reactive metabolic products, virus infections etc.) are added between generations. The theme of this article is copy fidelity so I will focus now on just this contribution.
We can use the analogy of a photocopier. In normal operation a photocopier can produce many copies of a single original document and all copies will be the same (if the machine is working correctly). To convert the photocopier into a biological analogue we need to take a copy of the original, then replace the original with its copy, and produce a ‘copy of a copy’. By replacing the ‘original’ each time with the copy of itself we create a ‘copy of a copy of a copy’ and so on, and this represents fairly closely what happens when organisms copy their genomes into their offspring.
We can represent this copying process mathematically with the following equation:
Q = PN (1)
where Q is the copy fidelity today, P is the post-Fall copy fidelity, and N is the number of generations since creation. This model is not quite correct in the sense that the value of P should be decaying at least to a small degree over time because ever since the Fall the whole creation has been ‘in bondage to decay’ (Romans 8:21). We can therefore introduce a decay term, α< 1, that can reduce the value a small amount each generation. The equation then becomes:
Q = αN × PN = (αP)N (2)
This model simulates the behaviour of an intelligently designed machine like the photocopier where its purpose is to make large numbers of high-quality copies over many years, and it receives regular maintenance and repair to keep it working correctly. But even with repair and maintenance no machine can last forever, so the value of α must decline slowly over time. As a first approximation we can make a decline at a similar rate to Q. This can be implemented by calculating a new value of α at the beginning of each new generation as follows:
α = Q = PN (3)
When this value is substituted into equation 2 we get:
Q = PN x PN = P2N (4)
Shortly we will be considering naturalistic Darwinian scenarios for the origin of life. In these cases there are no intelligently designed machines, and no maintenance or repair is available to anything that might happen to turn up by chance. To simulate this kind of process we need a model in which copy fidelity declines at the same rate as Q over time. In genomic terms this means every part of any primordial ‘genome’ would be as prone to copy error as any other part and the copy mechanism itself would degrade at the same rate as everything else. This model can be expressed mathematically as follows:
QN = (QN-1)2 (5)
where QN is the copy fidelity in generation N, QN-1 is copy fidelity in the previous generation, and QN = P in the beginning, when N = 1.
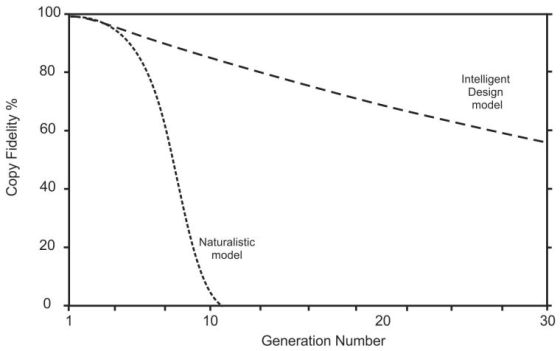
Figure 1 illustrates the difference in the way these models behave when they begin with a copy fidelity of just 99%. The Naturalistic model (equation 5) plummets to extinction very quickly, while the Intelligent Design (ID) model (equation 4) lasts much longer. As copy fidelity gets closer and closer to 100%, however, the output from both models will last for longer and longer times.
To execute these two models in practice we can adopt a trial-and-error approach.16 We begin by choosing a trial estimate of the original copy fidelity, then apply the different multiplication methods over 5,750 germ-cell generations and compare the result with the copy fidelity we observe today. Then we adjust the original number and try again, and repeat the process until we get the exact result observed today. I carried this out using both analytical solutions and a numerical routine written in Visual Basic within MS Excel.
Now we don’t know what the exact error rate today is from copy error alone. The Hutterite number can serve as a best guess because it is from a people group who live simple lives in rural colonies and have lower exposure to radiation, chemicals and other damaging factors that exist in high-tech communities. Their rate is also lower than the figures derived from other methods. So our target number for today’s copy fidelity will be 1.2 errors per 100 million nucleotides per human generation, which is equivalent to 1.2 ÷ 23 = 0.052 per 100 million, or approximately 1 error per 2 billion nucleotides per female germ-cell division.
A second target number that a successful model needs to match is the number of copy errors since creation, estimated earlier at 9,000 (36 per generation over 250 generations) in total. A successful model needs to produce a number less than 9,000 because copy errors must have been fewer in the past than they are today. And a third target for a successful model is that copy fidelity must continuously decline at a increasing rate as the copy mechanism becomes more and more degraded over time.
Modelling results
Using the Naturalistic model (equation 5) the copy fidelity declined rapidly. Even with a primordial error rate as low as 1 in ten thousand trillion (10 to the 16th power)—the smallest number that MS Excel would accept—the genome quality declined to extinction in just 55 germ-cell generations. This would be equivalent to the third human generation from Adam! In other words, copy fidelity could not possibly have declined at the same rate as genome quality or humans would already be extinct. The Author of life must have specially designed a system to protect the copy mechanism from errors!
The ID model (equation 4) worked much better. However, precision limits and rounding errors interfered severely in the MS Excel calculations so alternatives were required. I used the arbitrary precision function in Wolfram Mathematica v.9.0.1 to do Surd root solutions to equations 1 and 4 and for the numerical calculations at 50 and 100 digit precision. The ID model required a post-Fall copy fidelity of 1 error per 23 trillion nucleotides per germ-cell generation to fit the value of Q we observe today at 1 error in 2 billion nucleotides per germ-cell generation. Both high-precision analytical and numerical methods produced exactly this same result, but the MS Excel calculations produced values that differed by more than a thousand times!
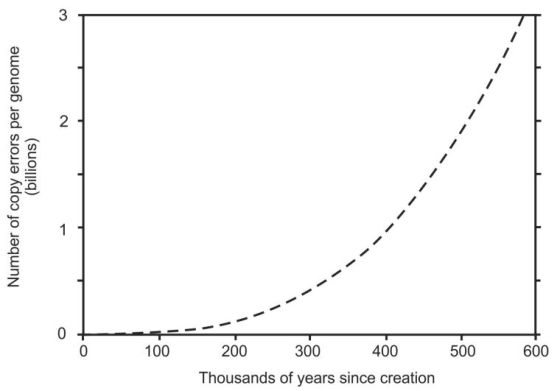
Two different values of α were tried: α = Q as given in equations 3 and 4, and α = 1 which is equivalent to equation 1 and implies a higher standard of ongoing repair and maintenance. Equation 4 in the high-precision numerical model yielded 3,834 to 4,313 mutations since creation (with and without rounding, respectively). Using α = 1 the primordial copy fidelity required was 1 error per 15 trillion nucleotides per germ-cell generation. When this value was used in equation 4 with no rounding, it yielded 3,307 mutations since creation. All these values are less than the 9,000 mutations calculated from the Hutterite error rate so they all meet that criterion successfully. Equation 4 with α = Q also produced a continuously increasing copy error rate, illustrated in figure 2, thus meeting the third criterion for a successful model. In contrast, using α = 1 failed this test as it became linear, producing exactly 1 million mutations every hundred thousand germ-cell generations.
The number of mutations per person per generation ranged from zero (perfect copy fidelity) during the first 3 human generations (4 human generations when α = 1), to the equivalent of every nucleotide being mutant from copy errors alone in less than 600,000 years (figure 2).
We don’t know how many mutations would cause extinction in humans, but since 99.9% of our mutations arise from causes other than copy errors (4,000 is only 0.13% of 3 billion) then extinction would occur very much sooner than figure 2 suggests. The only experimentally determined value available for mutational decay to extinction for any species is for the 1918 H1N1 influenza virus. Extinction occurred when about 10% of its genome was damaged.17 Ten percent of the human genome is 300 million mutations and that was achieved in the ID model after ~27,000 years.
Discussion of modelling results
The Naturalistic model is clearly incapable of matching the real genome data targets because it collapses so rapidly into mutation meltdown. In contrast, the ID model nicely fits all three data targets. Yet how can such an incredibly high mathematical precision be biologically possible? Have we made a false assumption somewhere? For example, does copy fidelity really propagate across generations in the multiplicative manner assumed here?
Genome copying occurs at a stage in the cell cycle called ‘DNA replication’. The double helix is unwound at multiple points on each chromosome and each exposed strand of the helix has new nucleotides attached to it by the copying machinery. The unwinding helix thus turns into two new double helices which are (almost) identical copies of the original. The two copies are then pulled apart at mitosis when the cell divides into two. The whole process is quite complex, involving many different molecular machines that include proofreading, error correction and error accumulation checkpoints. Overall, however, the occurrence of errors is fairly random, and thus a probabilistic process. Probabilities always propagate in a multiplicative manner because the chance of an error in one step is largely independent of the chance of error in the preceding or following step.
Natural selection also contributes towards maintaining high copy fidelity. When a female produces 7 million egg cells in the developing embryo and then selects just 1 or 2 million to store in her mature ovaries, her body is probably selecting the healthiest and rejecting those that carry the most mutations. Natural selection also plays a strong role in fertilization, ensuring that only the fittest one out of millions of sperm cells gets to fertilize the single egg that is produced in each reproductive cycle. Likewise, during embryo development, any that are badly damaged usually abort spontaneously so that only the relatively healthy babies make it to full term.
Another possibility here is that human female egg production may not be the same as in other mammals. The description given earlier was based on the generalised process in mammals but we need to realize that ethical issues limit the amount and type of research that can be done on humans.
The idea that all eggs are produced during embryogenesis has recently been challenged by the discovery that egg production can be artificially stimulated in cultures derived from cells scraped from the surface of adult human ovaries. This ability was described by the authors as a ‘sophisticated mechanism created during the evolution of female reproduction’18 and suggested that it might be a way of producing new high quality eggs when the embryonic store has become depleted. It will be interesting to see what future genomic studies of the human egg production process will reveal about mechanisms for conserving copy fidelity.
The large burden of mutations that the current human population carries, and passes on to its children, is not the result of copying errors—something else must have caused it.
Darwinian origin of life scenarios
With that background of real genome data and analysis in mind, we are now in a position to evaluate Darwinian scenarios for the origin of life. The Darwinian worldview requires life to have arisen through a long sequence of small, easy steps that could have occurred by chance and accumulated through natural selection to produce life as we know it today. No one knows how this might be possible. At least seven different scenarios have been put forward19 but despite intensive ongoing research20,21,22 and increasingly learned and detailed discussions, the same old ‘brick walls’ are encountered on every side.
That does not deter the true believers. For example, University of California, Berkeley, maintains a website called Understanding Evolution and under the topic “From Soup to Cells—the Origin of Life”23 they give no hint of any difficulty. On the contrary they say there is “illumination” from “many lines of evidence … even experiments”.
On the more focused topic “How did life originate?” they suggest that ribonucleic acid (RNA) formed spontaneously and began replicating itself. These self-replicators were then superseded by ‘super-replicators’ which took over via natural selection and these were then superseded by ‘supersuper-replicators’ which again took over and so on. Then, by chance, something even better turned up: self-replicating molecules became enclosed within a cell membrane. They state: “Cell membranes must have been so advantageous that these encased replicators quickly out-competed ‘naked’ replicators. This breakthrough would have given rise to an organism much like a modern bacterium.”
So, according to the Darwinian view, sloppy copying led to better sloppy copying, and sloppy copying that occurred inside a cell membrane led to better-than-ever sloppy copying.
But does it?
Self-replicating RNA molecules have been produced in the laboratory for medical and research purposes, but they require a great deal of intelligent design and manipulation.24 Evolutionists have long known that there are numerous obstacles to its naturalistic formation.25
A story in New Scientist entitled “Biologists create self-replicating RNA molecule” illustrates some of these problems.26 The molecule, dubbed tC19Z, could only replicate 97 nucleotides—not quite half its own length. The precursor to tC19Z, called R18, which the investigator began with, could only copy 14 RNA nucleotides, just 7% of its own length. The investigator made a vast library of thousands of different versions of R18 and screened them to see which ones made more copies. After many rounds of copying and screening he found several useful things which he incorporated into his final version. But even after all this intelligent design and manipulation, tC19Z could only copy 48% of its own length.
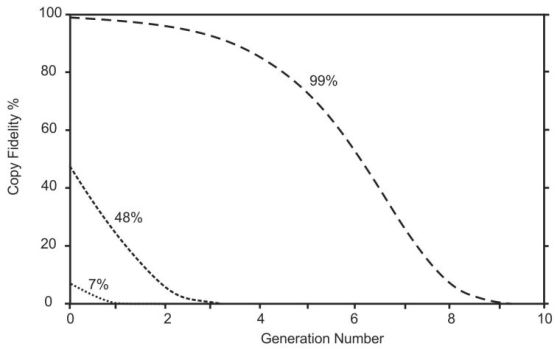
The foolishness of the claims made for these results—that they help us to understand how life could have arisen by chance—is illustrated in figure 3. In a primordial scenario like this only the Naturalistic model of copying could apply because the ID model requires special help and so is ruled out by definition. Suppose we begin with an RNA molecule that had a self-replicating capacity of 7% copy fidelity (like the R18 molecule) it would degenerate into useless junk after just 1 generation. With 48% fidelity (like the tC19Z molecule) there would be nothing useful left after 3 replications. Even if an experimenter managed to make an RNA molecule with 99% copy fidelity it would decay away in just 9 generations.
These results clearly show that anything other than extremely high copy fidelity quickly leads to error catastrophe! This result has been in the scientific literature for more than 40 years since Manfred Eigen coined the term ‘error catastrophe’ to describe exactly this problem.27 He calculated that only an error rate of less than 1/n where n is the effective genome size could sustain life. An average bacterial genome contains about a million nucleotides so the primordial error rate would need to be less than 1 in a million. Bacteria today can have in vivo mutation rates28 lower than 1 error per billion nucleotides per generation,29 a thousand times lower than this threshold and sufficient to keep them viable in the long term.
Clearly a great deal more intelligent design and manipulation than that which went into these RNA experiments would be required to create and sustain life over thousands of years!
General discussion and conclusions
The only kind of genome copying system that can sustain life over thousands of years is one that has two primary characteristics. First, it must be precisely engineered so that it can begin with copy fidelity so high as to be indistinguishable from perfection. Second, it must be protected and maintained over thousands of years in such a way that it is at least partly insulated from the general genome decay that is rapidly going on around it.
These characteristics fit very well into the biblical account of Creation and Fall just a few thousand years ago, but are impossible to achieve from a Darwinian starting point. The Darwinian claim—that life could have started with low-fidelity self-replicating molecules—is exposed as culpable foolishness. The ‘RNA world’ scenario collapses into ‘error catastrophe’ so quickly it is scandalous that such nonsense can be taught as a realistic model of origin in our schools and universities.
The ID model was not set up with the aim of producing perfect copy fidelity; it was simply projected backwards in time to a date consistent with biblical origins. The fact that it did produce perfect copy fidelity at around 4,000 BC—in both versions—is an independent result that exactly reflects our expectations from the Genesis account. The two values of α were chosen to represent two extremes. When α = 1 the standard of repair and maintenance does not decline over time (and is not to be expected in a fallen world), and when α = Q the standard of repair and maintenance declines at the maximum rate consistent with long-term survival and is a much more realistic scenario. Since both these models converge on perfect copy fidelity around 4,000 BC they leave no room for an earlier date for creation.
The copy fidelity in our cells today is a million times higher than the general genome quality, so it should remain in good shape and continue its work of high-fidelity copying for many generations to come. But it cannot last forever. Even if nothing else intervened it would come to an end through its own degeneration within thousands (not millions) of years. Meanwhile, about 99.9% of our mutations are coming from sources other than copy errors. It is these that will intervene and lead to our extinction via ‘mutation meltdown’ well before then. Human ancestors cannot have been around for more than a few thousand years or it would have already happened!
It will be an interesting challenge to identify likely causes of the 99.9% of mutations that come from non-copy-error sources. Accelerated nuclear decay is one possibility.30 Another interesting point is that because of the long lives of the early patriarchs, Abraham was just the 19th generation after the Fall and the ID model with α = Q predicted an average of just 25 copy errors per genome at that time. This may partly explain marriage practice at the time being preferred within families (Abraham, Isaac and Jacob all married close relatives). Only later, in the time of Moses, did God prohibit marriage between close relatives—when mutation burden would have begun to increase. This model only deals with copy errors, however, and at any time post-Fall a significant mutation burden could have, and clearly did, arise from other sources.
Acknowledgments
Three anonymous referees contributed greatly to the improvement of the original article. Bruce Axtens kindly provided advice on the precision limitations of MS Excel and possible alternative software and assisted with the high-precision numerical analyses.
References and notes
- Sanford, J.C., Genetic Entropy & the Mystery of the Genome, 3rd edn, FMS Publications, New York, 2008. Return to text.
- Williams, A.R., Mutations: evolution’s engine becomes evolution’s end, J. Creation 22(2):60–66, 2008; creation.com/evolutions-end. Return to text.
- Sanford, J.C. and Nelson, C.W., The Next Step in Understanding Population Dynamics: Comprehensive Numerical Simulation; in: Studies in Population Genetics, Fusté, M.C. (Ed.), InTech, 2012, intechopen.com, accessed 20 June 2013. Return to text.
- Kunkel, T.A., DNA Replication Fidelity, J. Biological Chemistry 279(17):16895–16898, 2004. Return to text.
- Fijalkowska, I.J., Schaaper, R.M. and Jonczyk, P., DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair, FEMS Microbiology Reviews 36(6):1105–1121, 2012. Return to text.
- Campbell, C.D. et al., Estimating the human mutation rate using autozygosity in a founder population, Nature Genetics 44:1277–1281, 2012 | doi:10.1038/ng.2418. Forty-four autozygous segments were found in which 72 heterozygous SNPs were validated within a total of 512 megabases of autozygous DNA. Return to text.
- An SNP (single nucleotide polymorphism), or SNV (single nucleotide variation) occurs when a base-pair is changed in relation to the corresponding position on a comparable chromosome (e.g maternal vs paternal, parent vs child, one genome vs. another genome). Return to text.
- Fu, W-Q., et al., Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants, Nature 493(7431):216–220, 2013 | doi:10.1038/nature11690. Return to text.
- One cell divides into two, two into four, etc. and after 23 such generations there are theoretically 8,388,608 cells (=2 multiplied by itself 23 times). Return to text.
- Oogenesis, en.wikipedia.org, accessed 20 June 2013. Return to text.
- The 1000 Genomes Project Consortium, An integrated map of genetic variation from 1,092 human genomes, Nature 491(7422):56–65, 2012 | doi:10.1038/nature11632. Return to text.
- Yi, X. et al., Sequencing of 50 human exomes reveals adaptation to high altitude, Science 329(5987):75–78, 2010 | doi:10.1126/science.1190371. Return to text.
- Rasmussen, M. et al., Ancient human genome sequence of an extinct Palaeo-Eskimo, Nature 463:757–762, 2010 | doi:10.1038/nature08835. Return to text.
- For simplicity, we are neglecting the genetic bottleneck resulting from the Flood. Most of the SNPs generated during the roughly 2000 preceding years would not have been carried by the eight Flood survivors. Return to text.
- Since human genomes are huge, we can make the reasonable assumption that 36 SNPs per generation are unlikely to produce duplicate mutations. Return to text.
- The exact solution requires a Surd function to calculate the 5,750th root of the square root of P, or the 11,500th root of P. Return to text.
- Carter, R.W. and Sanford, J.C., A new look at an old virus: patterns of mutation accumulation in the human H1N1 influenza virus since 1918, Theoretical Biology and Medical Modelling 9:42, 2012. Return to text.
- Bukovsky, A. et al., Oogenesis in cultures derived from adult human ovaries, Reproductive Biology and Endocrinology 3:17, 2005; rbej.com, accessed 13 August 2013. Return to text.
- Choi, C.Q., 7 Theories on the Origin of Life, Live Science, 22 March 2011, livescience.com, accessed 27 June 2013. Return to text.
- Powner, M.W. and Sutherland, J.D., Prebiotic chemistry: a new modus operandi, Philos Trans R Soc Lond B Biol Sci 366(1580):2870-7, 2011. | doi: 10.1098/rstb.2011.0134. Return to text.
- Szostak, J.W., The eightfold path to non-enzymatic RNA replication, J. Systems Chemistry 3:2, 2012. Return to text.
- Vasas, V. et al., Evolution before genes, Biology Direct 7:1, 2012. Return to text.
- How did life originate?, evolution.berkeley.edu, accessed 27 June 2013. Return to text.
- Ying, H. et al., Cancer therapy using a self-replicating RNA vaccine, Nature Medicine 5:823–827, 1999. Return to text.
- Evolutionist criticisms of the RNA World conjecture, June 2006, creation.com/rna. Return to text.
- Biologists create self-replicating RNA molecule, newscientist.com, accessed 2 July 2013. Return to text.
- Eigen, M., Self-organization of Matter and the Evolution of Biological Macromolecules, Max-Planck-Institut für Biophysikalische Chemie, Göttingen, 1971. Return to text.
- Measured in bacteria living in their natural environment rather than in a test tube. Return to text.
- Ford, C.B. et al., Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection, Nature Genetics 43:482–486, 2011 | doi:10.1038/ng.811. Return to text.
- De Young, D., Thousands … Not Billions: Challenging an Icon of Evolution, Questioning the Age of the Earth, Master Books, Green Forest, AR, chap. 9, 2005. Return to text.












Readers’ comments
Comments are automatically closed 14 days after publication.