Evidence that modern humans and ‘super-archaic’ humans may have shared DNA
Evolutionary views on human origins catch up with the Bible!

A recent paper in the journal Science suggests that the ancestors of modern humans, Neandertals, and Denisovans interbred with so-called ‘super-archaic’ humans on at least two separate occasions.1 Such super-ancient humans could possibly include Homo erectus, Homo heidelbergensis, or even some other group, according to some of the scientists examining the genetic material of modern humans.
If we reinterpret the data in this paper, we find that Neandertals, Denisovans, and now some unnamed super-archaic groups (possibly H. erectus) are not separate evolutionary lineages, but rather they are the same species. How can we do this? What did these researchers find?
Modern humans, Neandertals and Denisovans
As a first step, let’s examine how views on the relationship between Neandertals and modern humans has changed over the past few decades. About 150 years ago, scientists believed that Neandertals were a transitional form between apes and modern humans. Since then, researchers have done a complete 180. Virtually all scientists now accept that Neandertals and their distant kin, the Denisovans, discovered in 2008 in Siberia, are related to modern humans.
There are many fossils with Neandertal features in the fossil record. For example, a fossil from Jebel Irhoud in Morocco also shows mixed human and Neandertal features. Furthermore, a whole slew of Neandertal traits show that they shared several cognitive features with modern humans. These include the use of plants to make medicine, the production of composite tools, hunting patterns, trading routes,2 and diving for shellfish.3
Genetic evidence also shows that Neandertals and Denisovans are very similar to modern humans. The reason we know this is because researchers have extracted DNA from Neandertal and Denisovan bones, sequenced it, and assembled their genomes.4,5 The fact that researchers were able to do this means that Neanderthal and Denisovan bones aren’t hundreds of thousands of years old, because DNA is a fragile macromolecule which degrades very rapidly.
Genetic comparisons between humans and Neandertals show that Neandertals overlap the genetic variability of modern humans, with few fixed differences. Modern people outside of Africa inherited 2% or more of their DNA from Neandertals. If you analyze your ancestry using 23andMe.com, you can even get a report on how much of your DNA can be traced back to Neandertals.6 Denisovans share genes with modern humans involved in hemoglobin structure7 and the immune system. Tibetans seem to have inherited their ability to live at high altitudes from Denisovans. On average, 0.5% of Asian DNA comes from Denisovans, whereas Melanesians share up to 7% of their DNA with Denisovans. Also, methylation patterns are very similar between modern humans, Neandertals and Denisovans, and much more than they are to chimpanzees.8 Some secular scientists now call the ancestors of Neandertals and Denisovans by a new composite name, the ‘Neandersovans’.
According to the ‘biological species concept’,9 two organisms belong to the same species if they can interbreed. We can find evidence of interbreeding between two organisms if we find DNA shared between them. In summary, based on the fossil and genetic evidence, we can be certain that modern humans, Neandertals, and Denisovans are one species, and belong to the same created kind. In other words, they are descendants of Adam, descendants of Noah, and fully spiritual beings with a human mind and soul.
But what about the ‘super-archaic’ humans mentioned earlier? No DNA has been extracted for these sparse fossils, so how do researchers know that humans mixed with these groups as well? And are they more ‘archaic’ than the others?
Finding super-archaic humans
For the past several years, researchers have been hinting that they suspected modern humans received DNA from super-archaic humans. According to Murray Cox, a computational biologist from Massey University in New Zealand,
“It’s now clear that interbreeding between different groups of humans goes all the way back [to the beginning].”1
With the sequencing of a Neandertal genome from the Altai mountains in south central Russia in 2014, researchers found evidence of gene flow between this individual and Denisovans. This is nothing new, but the researchers also estimated that between 3–6% of the Denisovan genome itself came from an unknown archaic human group.10
To be certain that an alleged archaic group of humans interbred with modern humans, we need a copy of their actual genome sequence. This way, if we find pieces of ‘archaic’ DNA in the genomes of modern humans, we know that they interbred. Scientists have the genome sequence of only Neandertals and Denisovans. With the genome sequence lacking from other archaic humans, we can only infer the presence of archaic DNA in modern humans via statistical models. The results of these models are more tentative than simply comparing the genomes of two individuals. In fact, there is quite a bit of guesswork involved. Figure 1 summarizes models portraying gene flow between modern humans, Neandertals, Denisovans and super-archaic humans.
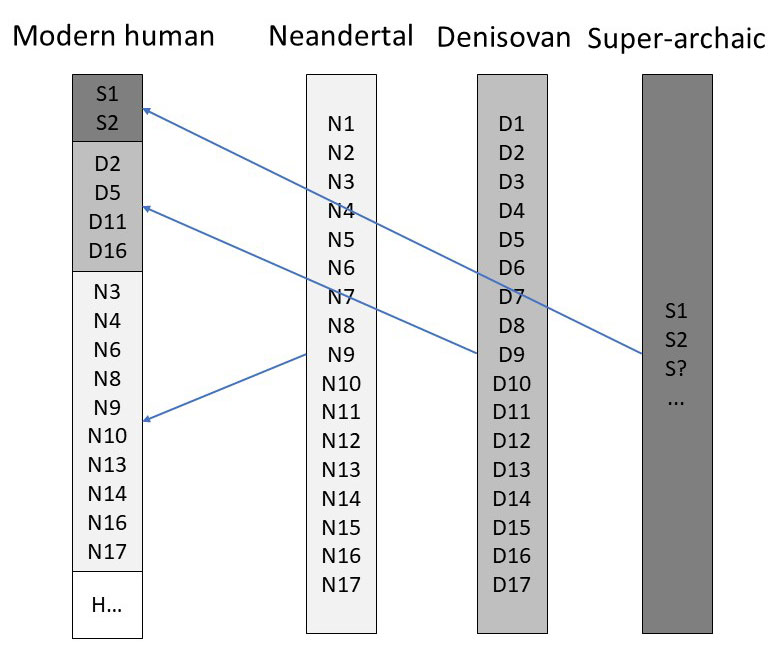
Researchers infer that a portion of DNA from modern humans comes from a super-archaic lineage when they find variants in the human genome that are missing from the genomes of both Neandertal and Denisovans (see figure 2). They also need to compare Africans to non-Africans, using the supposed out-of-Africa event as a time marker for introgression. This is what Durvasula and Sankararaman did when they compared the genomes of four sub-Saharan people groups with Neandertal and Denisovan. They estimated that between 2–19% of the genome of these supposed super-archaic groups, including a handful of genes which play a role in tumor suppression and hormone regulation, comes from a human lineage pre-dating Neanderthals and Denisovans.11 They based the conclusion on the fact that these variants are not found in Neanderthals and Denisovans. But, of course, they are assuming there was an earlier population. It is also possible that some genetic variants were passed to the Neanderthals, and not to Africans, and vice versa, from the early post-Flood population.
Besides genetic evidence that super-archaic humans interbred with humans, other studies involving physical characteristics show that archaic humans like H. erectus and H. heidelbergensis are related to modern humans.12,13,14
This is significant. Since there is evidence of genetic intermixing between modern humans, Neandertals, Denisovans, and now super-archaic groups, mean that humans form a single group, instead of multiple evolutionary lineages remaining independent of one another for millions of years. This is exactly what evolutionists are now coming to say based on fossil and ancient genomic evidence from all across Africa: humans have descended from a common ancestor, sharing many features, but also showing a lot of physical variation.15
Overturning evolutionary models of speciation
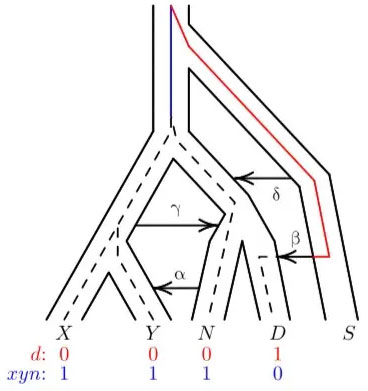
Besides this, there is another surprising conclusion that we could draw from these studies, if they are true. Evolutionists have tried to determine when and where the genetic mixing occurred between super-archaic humans, Neanderthals, Denisovans, and modern humans. The evolutionary models contradict one another, and they are forced to involve complex movements of archaic people groups back and forth between Africa and Eurasia.
One research group studied DNA letter patterns in Africans, Eurasians, Neandertals and Denisovans. They inferred that DNA admixture happened once between super-archaic humans and the ancestors of Neandertals and Denisovans and again between super-archaic humans and Denisovans (figure 3).15 Even though they did not have the super-archaic genome available, they still modelled when gene flow happened between which genomes, based on the pattern of DNA letters at specified locations in the genomes of Africans, Eurasians, Neandertals and Denisovans. This is very speculative work. It also assumes deep time and common ancestry with chimps. In fact, the ‘ancestral’ state was determined by comparing to chimpanzee DNA. If this assumption is incorrect, the evidence of mixing with some early-post-Flood ‘super-archaic’ humans might stand, but the timing and order of events will be radically different.
Yet, the idea that super-archaic humans could have interbred with Neandertals, Denisovans, and modern humans has overturned a long-standing component of evolutionary theory. According to Pontus Skoglund of the Francis Crick Institute in London,
“We are losing the idea that separation between populations is simple with instant isolation.”1
According to evolutionary theorists, when a group of organisms divides into two sub-groups, they will, eventually, no longer be able to breed with one another. This is because when the two sub-groups are isolated, an increasing number of mutations accumulate in their respective genomes, eventually making them genetically incompatible with one another.
This pattern of mixing after divergence is quite common in nature. But it is entirely surprising for the evolutionist and they have been very slow to acknowledge it.16 At least 10% of animal species, and 25% of plant species are capable of breeding with other plant or animal species.17 For example, gene flow has been discovered between the Asian black bear, and the ancestor of the polar, brown, and American black bear.18 This is proof that bears also belong to one kind.
Why does this matter?
Earlier, many evolutionists believed different human groups had separate origins, each spanning millions of years. However, based on new genetic evidence, some evolutionists have revised their views on the relationships between modern humans, Neandertals, Denisovans, and super-archaic humans. They now freely admit that there could have been significant gene flow between these groups. If true, then this means that they must be inter-related, forming one species, with one origin. In the end, all these groups would be human, descended from Adam and the people who came off the Ark.
This is what the Bible has been saying all along. It is quite remarkable that evolutionists could finally come to this same conclusion (that modern and archaic humans are all related). It means that Christians don’t have to be afraid of every scientific theory that seems to contradict the Bible, because the evidence ultimately fits in with the biblical narrative—it means you can trust God’s Word.
References and notes
- Gibbons, A., Strange bedfellows for human ancestors, Science 367(6480):838–839, 2020. Return to text.
- O’Micks, J., Modern science catches up with Neandertal man, J Creation 32(1):38–42, 2018. Return to text.
- Stringer, C.B. et al., Neanderthal exploitation of marine mammals in Gibraltar, PNAS 105(38):14319–24, 2008. Return to text.
- A genome is the entirety of an organism’s DNA, all of its genes and chromosomes. Return to text.
- Cserhati, M.F. et al. Motifome comparison between modern human, Neanderthal and Denisovan, BMC Genomics 19(1):472, 2018. Return to text.
- Neanderthal Ancestry Report Basics, customercare.23andme.com/hc/en-us/articles/212873707-Neanderthal-Ancestry-Report-Basics, accessed 6 March 2020. Return to text.
- A protein found in red blood cells which carry oxygen to our cells. Return to text.
- Gokhman, D. et al., Reconstructing the DNA methylation maps of the Neandertal and the Denisovan, Science 344(6183):523–527, 2014. Return to text.
- de Queiroz K., Ernst Mayr and the modern concept of species, Proc Natl Acad Sci U S A, 102 Suppl 1:6600–7, 2005. Return to text.
- Prüfer K. et al., The complete genome sequence of a Neanderthal from the Altai Mountains, Nature 505(7481):43–9, 2014. Return to text.
- Durvasula, A. and Sankararaman, S., Recovering signals of ghost archaic introgression in African populations, Sci Adv. 6(7): eaax5097, 2019. Return to text.
- O’Micks, J., Rebuttal to “Reply to O’Micks Concerning the Geology and Taphonomy of the Homo naledi Site” and “Identifying Humans in the Fossil Record: A Further Response to O’Micks”, ARJ 10(2017):63–70. Return to text.
- O’Micks, Preliminary Baraminological Analysis of Homo naledi and Its Place Within the Human Baramin, Journal of Creation Theology and Science Series B: Life Sciences 6:31–39, 2016. Return to text.
- Lawton, G., Human evolution: the astounding new story of the origin of our species, New Scientist, newscientist.com/article/mg24532760-800-human-evolution-the-astounding-new-story-of-the-origin-of-our-species, accessed 21 April 2020. Return to text.
- Rogers, A.R., Legofit: estimating population history from genetic data, BMC Bioinformatics 20(1):526, 2019. Return to text.
- Pennisi, E., Shaking up the Tree of Life, Science 354(6314):817–821, 2016. Return to text.
- Mallet J., Hybridization as an invasion of the genome, Trends Ecol Evol. 20(5):229–37, 2005. Return to text.
- Kumar, V. et al., The evolutionary history of bears is characterized by gene flow across species, Sci Rep. 7:46487, 2017. Return to text.





Readers’ comments
Comments are automatically closed 14 days after publication.