Journal of Creation 33(2):51–56, August 2019
Browse our latest digital issue Subscribe
A successful decade for Mendel’s Accountant
A powerful computer program with far-reaching consequences has been developed by a group of biologists and computer scientists. Striking at the heart of neo-Darwinian theory, it tackles the subject of mutation/selection using a straightforward method called genetic accounting. Named Mendel’s Accountant, this software platform provides a comprehensive refutation of multiple aspects of evolutionary theory using nothing but standard evolutionary population genetics. The developers have used it to quantify the actual selection threshold for new mutations, to test alternate evolutionary ideas (e.g. unusual selection models, such as ‘synergistic epistasis’), to quantify the long-standing ‘waiting time problem’ for new beneficial mutations, to make predictions about the long-term effects of mutation accumulation in viruses (which were later confirmed), and to compare different historical population models to the modern human mutation frequency spectrum seen in the 1000 Genomes Project data. Their results represent a complete refutation of the ‘primary axiom’ of neo-Darwinian theory. Computationally, the mutation/selection model failed in multiple ways.
After the success of the Radioisotopes and the Age of the Earth (RATE) project,1 the Institute for Creation Research began another major endeavour they called the GENE project. Even though this second project received much less fanfare, it achieved significant results and documented these in multiple publications. Essentially, every major goal they began with has been achieved. My involvement in the GENE project led to the publication of the ‘mitochondrial Eve’ consensus sequence2 in Nucleic Acids Research and a follow-up paper at the 2008 International Conference on Creationism.3 But one aspect of the GENE project deserves special attention: the population-modelling program Mendel’s Accountant. This is a computer platform that enables users to test fundamental aspects of neo-Darwinian theory, using nothing but the tools of standard evolutionary population genetics. Major discoveries and advances were made possible thanks to the development of this powerful research tool.
Setting up the problem
For many years, neo-Darwinian population geneticists have been boasting about how successful their ideas were in making predictions, but they were obscuring significant problems. Specifically, they had grave mathematical difficulties when trying to explain how mutation and natural selection could produce the observed complexity of life. John Sanford calls mutation/selection the ‘primary axiom’ of neo-Darwinism in his groundbreaking book Genetic Entropy and the Mystery of the Genome.4 In it, he says that population geneticists know that most mutations are deleterious. Also, it is widely known that natural selection and the occasional beneficial mutation cannot stop the gradual decline caused by the accumulation of these slightly deleterious mutations. Individually, most mutations do no harm to the organism. But collectively, they can be catastrophic. They relentlessly accumulate in the genome over time because they are too weak to affect reproductive success. Since these deleterious mutations accumulate faster than they can be eliminated, the net result is genetic deterioration. When too many errors occur in a population, it enters a phase called ‘mutational meltdown’, which rapidly leads to extinction.
That is not merely a theoretical possibility. Given the measured rates of mutation and reproduction, Mendel’s Accountant shows that current evolutionary models result in genetic deterioration, suggesting that all complex life5 is degenerating. This fundamental problem exposes the unreality of Richard Dawkins’ popular (but misleading) computer program that generated the phrase ‘Methinks it is like a weasel’.6 Dawkins’ simulation used an unrealistically low rate of mutation (merely one per progeny), plus an unrealistically high rate of reproduction (approximately 200 progeny per female), plus an unrealistically high rate of beneficial mutation (one in 27), and it locked any ‘correct’ mutation in a step-wise fashion, as if each step in the right direction was more grammatically correct than the nonsense phrase that preceded it. In these ways, and more, Dawkins’ simulation was exceedingly unrealistic in favour of evolution.
The logic behind the genetic entropy problem was compelling, and was consistent with earlier mathematical analyses,7 but it was largely theoretical in nature when Sanford first wrote Genetic Entropy. Empirical testing of genetic entropy theory only became possible when a team of computer scientists and geneticists developed a computer simulation that could realistically and comprehensively model the mutation/selection process. The result was Mendel’s Accountant.8 This program models the mutation/selection process based upon the conventional, textbook understanding of evolutionary genetics. Unlike other modelling programs of this type, the key parameters are adjustable—allowing the simulation and testing of nearly any scenario of interest.
The program has undergone multiple updates and improvements, and significant new features have been added over the years (such as dynamic population size modelling). Since population modelling uses tremendous amounts of computer memory, the latest version is designed to run on large servers.
How does Mendel’s Accountant work?
Programs of this type need to be documented and validated in the professional literature. This happened in 2007 and 2008.9,10,11 The program tracks all the mutations that occur among the individuals in a modelled population using a process the authors call ‘genetic accounting’. When a simulated mutation occurs, it is assigned an identifier, a location on a chromosome, and a ‘fitness’ score using values that come from standard evolutionary genetics theory (i.e. although rare beneficial mutations do occur, almost all mutations are ‘slightly deleterious’). By allowing for chromosomal recombination, the accumulating mutations get mixed into various combinations along each chromosome. Since children randomly inherit half of their father’s and half of their mother’s genome, some mutations are immediately lost. When it comes time to reproduce, some individuals fail to reproduce because they carry a greater load of harmful mutations. These are the individuals that are selectively eliminated, similar to the way Charles Darwin imagined natural selection to work. The program is extremely flexible, by design. Various models of selection, fitness, mutation, mating, and population structure can be applied to a given simulation.
What have we learned from Mendel’s Accountant?
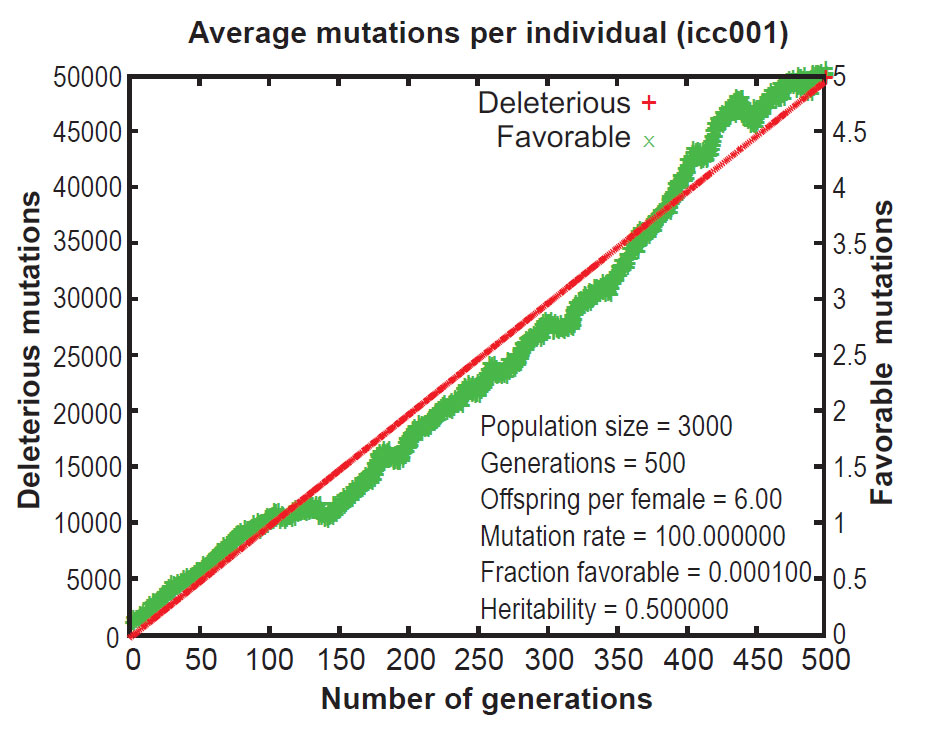
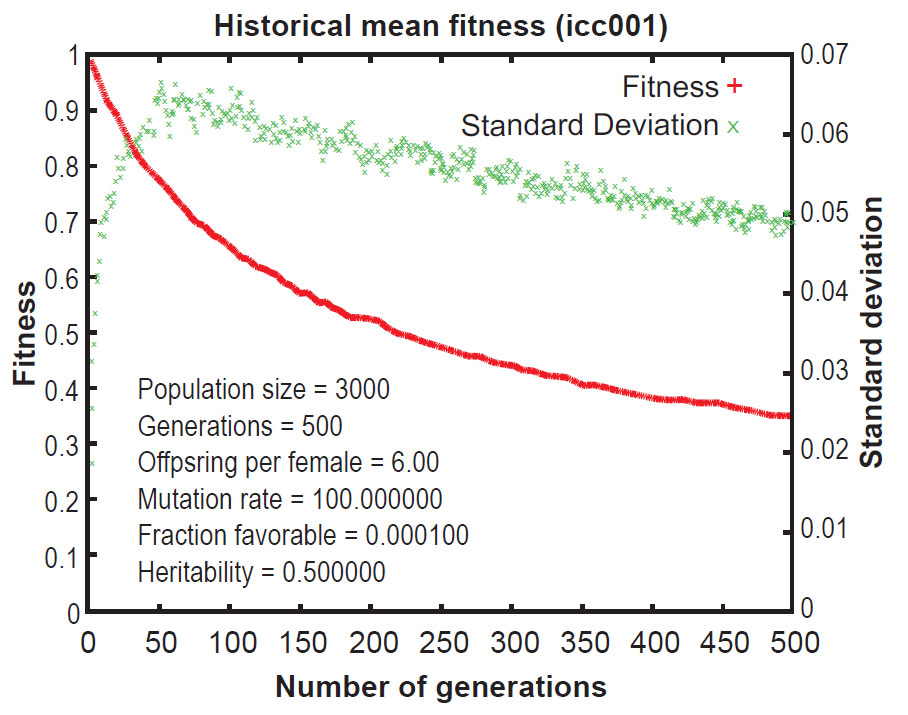
Early publications on Mendel’s Accountant showed that, under normal conditions, mutations accumulate in a highly linear manner. This results in a continuous increase in the number of mutations per progeny over time. This increase is inexorable and irreversible, even when selection pressure is strong. As a result of the ever-increasing mutational burden, the Mendel simulations revealed that ‘fitness’ (a measure of total functionality) declines continuously over time, even with strong selection (figures 1 and 2).12 This is a central problem for evolutionary theory, because if deleterious mutations cannot be removed, a species will deteriorate, all the way to the point of extinction. This is the essence of ‘genetic entropy’, and thus Mendel validates genetic entropy theory.
For the first time ever, we can now model how quickly mutations accumulate in large populations, the effects of natural selection, the effects of population bottlenecks, and what happens when rare ‘beneficial’ mutations appear, all thanks to this evolutionary modelling program.
A Mendel review paper appeared in 2012 as a chapter in an academic compilation of papers relating to population genetics.13 This chapter not only described how Mendel worked, but it also described how it compared to other significant evolutionary modelling programs. As of the time of writing, it was the only biologically realistic program of its kind. This is still true today.
Mendel’s Accountant is the most accurate software available for realistically simulating evolutionary genetic models. That has been true since its beginnings a decade ago. Why weren’t evolutionists the ones to produce it, or improve on it? Evolutionists had the know-how, plus access to super computers, and public funding. So why did a group of what they consider outsiders produce such a program and publicly promote it?
The answer is illuminating. There existed primitive forerunners of the Mendel’s Accountant simulation. These forerunners, created by evolutionists, were simple by comparison, yet when realistic values for human reproduction rate and mutation rate were used, they were already demonstrating genetic deterioration. In other words, the problem of genetic deterioration was known long ago, both in theory and in computer simulations, but they failed to pursue it further, as least not publicly.
Even today, anyone might attempt to refute the results of Mendel’s Accountant by producing a more realistic simulation. But they have not done so, not even in 10 years. This lack of serious response further suggests that Mendel’s Accountant is demonstrating real problems for evolutionary theory.
Biological Information—New Perspectives
In 2011, a conference was held at Cornell University entitled Biological Information—New Perspectives (BINP). Multiple speakers from a wide range of backgrounds presented papers at this conference, and Mendel’s Accountant was featured heavily. Indeed, one of the goals of the conference was to highlight its power, and six papers based on Mendel were delivered. Each paper is readily accessible online (see references).
A paper by Gibson et al. addressed the question of whether or not purifying natural selection could preserve biological information.14 They concluded that selection was unable to preserve genetic information15 because it systematically degenerates. Therefore, all genomes decay over time. Intense selection easily removed the worst mutations, but the mutations that were slightly deleterious accumulated relentlessly and without limit.
Sanford et al. empirically measured the threshold where selection breaks down (called the ‘selection threshold’) and determined what factors affect it the most.16 In other words, they measured how impactful a mutation must be before natural selection can ‘see’ it. They found that almost all mutations (both beneficial and deleterious) sit below the selection threshold, and so most mutations accumulate as if no selection was happening at all. When a mutational effect is below the selection threshold, that mutation is ‘effectively neutral’. It is unselectable. Its effect cannot rise above the genetic and environmental ‘noise’. This is true regardless of whether it is beneficial or harmful to the organism. None of the realistic simulations resulted in fitness gain.
Nelson and Sanford addressed the question of whether or not selection could preserve biological information using an alternative simulation.17 To do so, they rigorously compared results from Mendel experiments with results using one of the most popular evolutionary modelling programs out there, Avida. Interestingly, they discovered that an Avida-like model could only lead to evolutionary advance when extremely unrealistic parameter settings were employed in Mendel. Avida is highly unrealistic biologically. For example, Avida normally employs mutational effects that are grossly unrealistic (e.g. every step forward doubles total functionality/fitness). However, when realistic mutational effects were employed, there was no increase in fitness. Selection fails to create any new information, and any information already present degenerates toward ‘zero’, as shown by their detailed analysis of the inner workings of Avida.
Since the pioneering work of Kimura18 and Ohno19 in the late 1960s and early 1970s, evolutionary theory has recognized that, while most mutations are deleterious, individually their harmful effects are too subtle to be influenced by natural selection, and so cannot be effectively eliminated. Thus, a huge number of these ‘nearly neutral’ deleterious mutations accumulate continuously—like rust on a car. Even though an individual speck of rust is inconsequential to the function of an automobile, accumulating rust specks will eventually destroy it. Thus, mutations must be removed or the species will go extinct, and yet there does not appear to be any effective mechanism that can do this.
Two escape mechanisms have been proposed that, some have argued, might solve this problem. They are the ‘mutation count’ mechanism and ‘synergistic epistasis’. These two mechanisms are similar. They aim to make the selection process more efficient, but in unrealistic ways. The mutation count mechanism works by (unrealistically) counting mutations in each individual, ranking the individuals by that count, then eliminating only those individuals above a certain count threshold. Nature does not operate that way.
Brewer et al. analyzed the mutation count mechanism.20 They showed that it does not work, except when using extremely unrealistic parameter settings (such as forcing all mutations to have the same fitness effect). Whenever parameter settings were used that were even remotely realistic, the mechanism failed completely.
Synergistic epistasis assumes an (unrealistic) fitness model for describing how multiple mutations combine together in their effect on fitness. The idea is that when mutations interact with one another, they might sometimes amplify each other’s deleterious effects. Selection might then become more effective and the mutations could be removed simultaneously (essentially killing two or more birds with the same stone). This is different from the traditional view of epistasis. For example, when many genes work together to create a light-sensing cell in the retina, these genes have an effect greater than the sum of their individual effect. In other words, the whole eye is greater than the sum of its parts. Likewise, harmful mutations in these genes, in combination, have a greater harmful effect than the mere sum of their harmful effects (a few little changes can have catastrophic downstream effects). That idea is well-established and is traditionally known as ‘epistasis’. But ‘synergistic epistasis’ is quite different. It assumes it makes no difference whether the mutations are combined toward some useful combined function, such as sensing light. Instead it assumes any mutations, no matter what they affect, will have a magnified effect in any arbitrary combination. For example, it assumes harmful mutations to a toenail, the retina, and a tendon in the shoulder will have a magnified effect in combination, just as though they were doing something important together. This assumption is unrealistic. So unrealistic that evolutionary geneticists generally avoid it altogether (except when trying to solve such problems as genetic deterioration).
Baumgardner et al. examined synergistic epistasis.21 They conclusively showed that synergistic epistasis does not help stop deleterious mutation accumulation. In fact, it greatly accelerates genetic degeneration and leads to rapid extinction.
Bringing it full circle
There was one other paper presented at the BINP meeting that is important for this discussion. Brewer et al. raised an interesting question in their paper on mutation accumulation in RNA viruses, which they modelled using Mendel.22 Given the error rate inherent in RNA viral replication and the size of the viral genome, RNA viruses are prime candidates for genetic entropy.
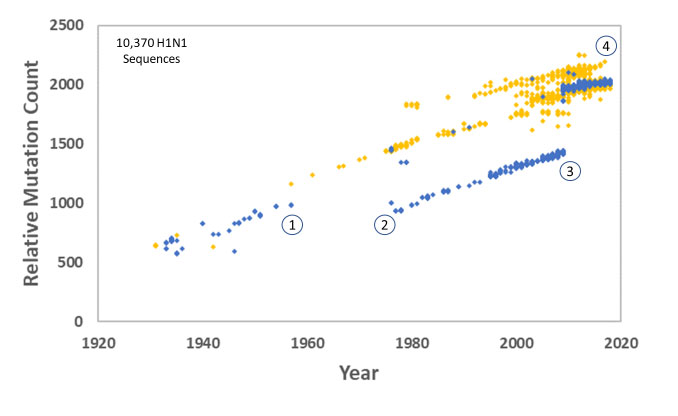
Carter and Sanford followed this up in 2012 with a study on the human H1N1 influenza virus, one of the most infamous RNA viruses in history, and the cause of the 1917/18 influenza pandemic that killed more people than died in WW1. We showed that mutation accumulation (despite lots of natural selection) led to the decline and eventual extinction of the virus (figure 3).23,24,25
This was the first time that genetic entropy theory was experimentally demonstrated in a biological system, completing the cycle from theory, to simulation, to application in the real world.
Ratcheting up the pressure
This flurry of papers from the Cornell symposium on biological information was not the end of the discoveries being made by Mendel. Rupe and Sanford wrote a fascinating paper that described a new principle they called ‘Haldane’s Ratchet’.26 They set out to demonstrate that the famous theoretical problem known as ‘Haldane’s Dilemma’ was indeed a serious challenge to evolutionary theory.27 Haldane’s Dilemma derives from theoretical work performed in the 1950s, where Haldane showed that, even when there are a sufficient number of beneficial mutations in a population, it takes too much time for selection to amplify them to the point where all individuals carry the ‘good’ mutations. The amount of time involved is simply unrealistic.
Haldane estimated that it would take 300 generations for a beneficial substitution to become ‘fixed’ (as in ‘stuck’, the point where all individuals carried a given mutation) in most populations. Thus, in three million years (the then-assumed time required for ape-to-man evolution), a pre-human population could only ‘fix’ about 500 beneficial mutations. This is a trivial amount of genetic information, which means Haldane’s Dilemma makes human evolution impossible. Rupe and Sanford used Mendel to simulate and experimentally validate that Haldane’s Dilemma is very real. In addition, they showed that net fitness declined due to the accumulation of deleterious mutations while the beneficial mutations were going toward fixation. In other words, for every beneficial mutation that was selectively fixed, a large number of nearly neutral deleterious mutations simultaneously drifted to fixation. The result is that, even in the most generous scenarios, net fitness only went down—it never went up. This one-way process acts like a ratchet, hence the title Haldane’s Ratchet, which is a hat-tip to another evolutionary paradox, Muller’s Ratchet,28 that deals specifically with mutation accumulation in asexual species.
In 2015, Sanford and colleagues published on the ‘waiting time problem’.29 Evolutionists already knew that in all modest-sized populations there is a long waiting time before any specific point mutation will arise. This greatly amplifies the problem of Haldane’s Dilemma, which assumes that all required beneficial mutations are already in the population.30 But single point mutations do not generally create new functions. For decades it has been argued how many beneficial mutations are needed, and how much time is needed, to typically evolve a new function. In this paper, they used Mendel to model strings of mutations in a human-like population. They showed that even a string of just two mutations required roughly 84 million years to arise and become fixed in the population. Of course, the string will appear more often than that, but since most mutations are lost to drift,26 even when beneficial, the real waiting time is much longer than the ‘first appearance’ time. A string of five point mutations would take approximately two billion years to arise, catch hold, and go to fixation! This problem is similar to Haldane’s Ratchet—but more than a million-fold worse. The authors modelled different mutation rates, fitness benefits, and population sizes, but even when using extremely generous settings, the waiting time problem is devastating for neo-Darwinian theory, especially for species with long generation times (like humans).
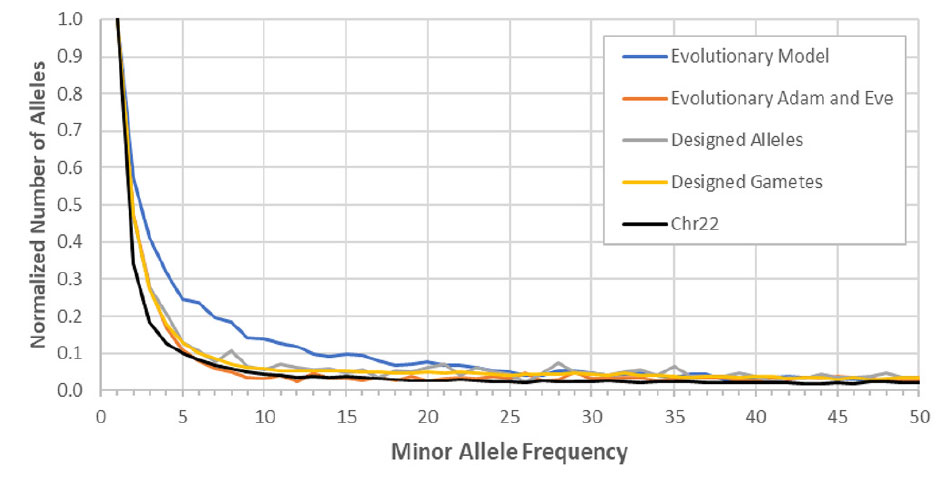
Finally, Mendel was used to show that simulations that employed alternative population models created allele distributions that better matched the actual allele frequency distribution (as seen in the modern human population) than did simulations that employed evolutionary parameters (figure 4).31 These results are intriguing. Since Mendel has already been demonstrated to be behaving according to standard evolutionary theory, the fact that alternatives to standard evolutionary theory better fit the real-world data should make us question some of the assumptions behind the ‘Out of Africa’ theory, for example.
Success has brought criticism
A Google search for Mendel’s Accountant will bring up multiple hits that criticize the program and the conclusions being drawn from its discoveries. Essentially none of these attacks are substantive, and many are highly misleading. It is clear that most people commenting on Mendel have not read either the documentation or the background papers. Thus, many evolutionists arguing against it don’t seem to understand their own theory. This is probably due to the anonymous nature of the internet, and the level of expertise required to make comments online (i.e. no understanding required). However, one anti-creationist blogger (not trained in either biology or genetics, and it shows) tried to build a credible case against the genetic entropy thesis, and thus tangentially attacking Mendel’s Accountant. John Sanford has rebutted that review in an important article on creation.com.32 We are unaware of any peer-reviewed paper that attempts to refute the methods or conclusions of Mendel. After a decade of established work, there should be something. Their silence is telling.
Conclusions
Mendel’s Accountant represents a milestone in our understanding of how the mutation/selection process operates. It shows what mutation/selection can and cannot do. For the first time, a robust computer program is able to successfully model realistic populations, track mutations through time, and rigorously test different evolutionary scenarios. No longer can evolutionists hide behind the mutation-count mechanism or the synergistic epistasis mechanism. No longer can they use selection as a ‘magic wand’ to wish away the theoretical, mathematical, or computational difficulties they face. Mendel is a powerful tool and should be used often. The collapse of the Darwinian mutation/selection mechanism is profound, and this news deserves more attention.
References and notes
- For one of many important publications that came from the RATE project, see Humphreys, R., Argon from RATE site confirms the earth is young; a second noble gas testifies to the biblical 6,000 years, Creation 34(3):40–42, 2012. Return to text.
- Carter, R.W., Mitochondrial diversity within modern human populations, Nucleic Acids Res. 35(9):3039–3045, 2007. Return to text.
- Carter, R., Criswell, D., and Sanford, J., The ‘Eve’ mitochondrial consensus sequence; in: Snelling, A.A. (Ed.), Proceedings of the Sixth International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, and Institute for Creation Research, Dallas, TX, pp. 111–116, 2008. Return to text.
- Truman, R., From ape to man via genetic meltdown: a theory in crisis; a review of Genetic Entropy & The Mystery of the Genome by John C. Sanford, J. Creation 21(1):43–47, 2007. Return to text.
- See Carter, R., Genetic entropy and simple organisms; if genetic entropy is true, why do bacteria still exist? genetic-entropy-and-simple-organisms, 25 October 2012. Return to text.
- Ey, L. and Batten, D., Weasel, a flexible program for investigating deterministic computer ‘demonstrations’ of evolution, J. Creation 16(2):84–88, 2002. Return to text.
- ReMine, W.J., Cost theory and the cost of substitution—a clarification, J. Creation19(1):113–125, 2005; creation.com/cost. Return to text.
- The software can be accessed at mendelsaccount.sourceforge.net. Return to text.
- Sanford, J., Baumgardner, J., Brewer, W., Gibson, P., and ReMine, W., Mendel’s Accountant: a biologically realistic forward-time population genetics program, SCPE 8(2):147–165, 2007. Return to text.
- Sanford, J., Baumgardner, J., Brewer, W., Gibson, P., and ReMine, W., Using computer simulation to understand mutation accumulation dynamics and genetic load; in: Shi, Y. et al. (Eds.), ICCS 2007, Part II, LNCS 4488, 2007. Return to text.
- Baumgardner, J., Sanford, J., Brewer, W., Gibson, P., and, ReMine, W., Mendel’s Accountant: a new population genetics simulation tool for studying mutation and natural selection; in: Snelling, A.A. (Ed.), Proceedings of the Sixth International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, and Institute for Creation Research, Dallas, TX, pp. 87–98, 2008. Return to text.
- Sanford, J., Baumgardner, J., Brewer, W., Gibson, P., and ReMine, W., Using numerical simulation to test the validity of neo-Darwinian theory; in: Snelling, A.A. (Ed.), Proceedings of the Sixth International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, and Institute for Creation Research, Dallas, TX, pp. 165–175, 2008. Return to text.
- Sanford, J. and Nelson, C.W., The next step in understanding population dynamics: comprehensive numerical simulation; in: Carmen Fusté, M. (Ed.), Studies in Population Genetics, InTech, chap. 7, pp. 117–135, 2012. Return to text.
- Brewer, W., Baumgardner, J., Gibson, P., and Sanford, J., Can purifying natural selection preserve biological information? in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds), Biological Information—New Perspectives, World Scientific, Singapore, pp. 232–263, 2013. Return to text.
- Carter, R.W., Can mutations create new information? J. Creation 25(2):92–98, 2011. Return to text.
- Sanford, J., Baumgardner, J., and Brewer, W., Selection threshold severely constrains capture of beneficial mutations; in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds.), Biological Information—New Perspectives, World Scientific, Singapore, pp. 264–297, 2013. Return to text.
- Nelson, C.W. and Sanford, J.C., Computational evolution experiments reveal a net loss of genetic information despite selection; in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds.) Biological Information—New Perspectives, World Scientific, Singapore, pp. 338–368, 2013. Return to text.
- Kimura, M., Evolution rate at the molecular level, Nature 217:624–626, 1968. Return to text.
- Ohno, S., So much ‘junk’ DNA in our genome—evolution of genetic systems, in: Smith, H.H. (Ed.), Brookhaven Symposia in Biology, no. 23, pp. 366–370, 1972. Return to text.
- Brewer, W., Baumgardner, J., and Sanford, J., Using numerical simulation to test the ‘Mutation-Count’ hypothesis; in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds.), Biological Information—New Perspectives, World Scientific, Singapore, pp. 298–311, 2013. Return to text.
- Baumgardner, J., Brewer, W., and Sanford, J., Can synergistic epistasis halt mutation accumulation? Results from numerical simulation; in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds.), Biological Information—New Perspectives, World Scientific, Singapore, pp. 312–337, 2013. Return to text.
- Brewer, W., Smith, F.D., and Sanford, J.C., Information loss: potential for accelerating natural genetic attenuation of RNA viruses; in: Marks II, R.J., Behe, M.J., Dembski, W.A., Gordon, B., and Sanford, J.C. (Eds.), Biological Information—New Perspectives, World Scientific, Singapore, pp. 369–384, 2013. Return to text.
- Carter, R. and Sanford J.C., A new look at an old virus: patterns of mutation accumulation in the human H1N1 influenza virus since 1918, Theor. Biol. Med. Model. 9:42, 2012. Return to text.
- Carter, R., More evidence for the reality of genetic entropy, J. Creation 28(1):16–17, 2014. Return to text.
- Carter, R., More evidence for the reality of genetic entropy—update, J. Creation 33(1):3–4, 2019. Return to text.
- Rupe, C.L. and Sanford, J.C., Using numerical simulation to better understand fixation rates, and establishment of a new principle: Haldane’s Ratchet; in: Horsetmeyer, M. (Ed.), Proceedings of the Seventh International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, 2013. Return to text.
- Batten, D., Haldane’s Dilemma has not been solved, J. Creation 19(1):20–21, 2005. Return to text.
- See the discussion on Mueller’s Ratchet in William, A., Healthy genomes require recent creation, J. Creation 29(2):70–77, 2015. Return to text.
- Sanford, J., Brewer, W., Smith, F., and Baumgardner, J., The waiting time problem in a model hominin population, Theoretical Biology and Medical Modelling 12:18, 2015. Return to text.
- Readers might also be interested in Basener, W.F. and Sanford, J.C., The fundamental theorem of natural selection with mutations, J. Mathematical Biology 76(7):1589–1622, 2018. Return to text.
- Sanford, J., Carter, R., Brewer, W., Baumgardner, J., Potter, B., and Potter, J., Adam, Eve, designed diversity, and allele frequencies; in: Whitmore, J.H. (Ed.), Proceedings of the Eighth International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, pp. 200–216, 2018. Return to text.
- Sanford, J., Critic ignores reality of genetic entropy: the author of a landmark book on genomic decay responds to unsustainable criticisms, creation.com/genetic-entropy, 7 March 2013. Return to text.








Readers’ comments
Comments are automatically closed 14 days after publication.