A family tree for all humanity?
Matthew H. from the USA wrote to us asking about a new paper that claims to have built a family tree for all the people in the world.

Good morning, I am a bible-believing Christian, and I accept a literal 6-day creation and an “age of earth” consistent with biblical genealogy (6k years). This morning, I came across an article describing a new “family tree” based on secular research. The tree is described as a draft; however, the age of the tree is over 100,000 years old. Obviously, this conflicts with the Bible’s timeline. I agree we should “Let God be true, and every man a liar”. Unfortunately, this is not a good answer to the unbeliever. I have searched your website for a good answer, but I haven’t had much luck. The research seems to be based on computer modeling, so I assume there are assumptions in the algorithms … Can you explain the assumptions? Please help me understand! I copied a link below for your convenience. [link deleted as per feedback rules]
Hi Matthew,
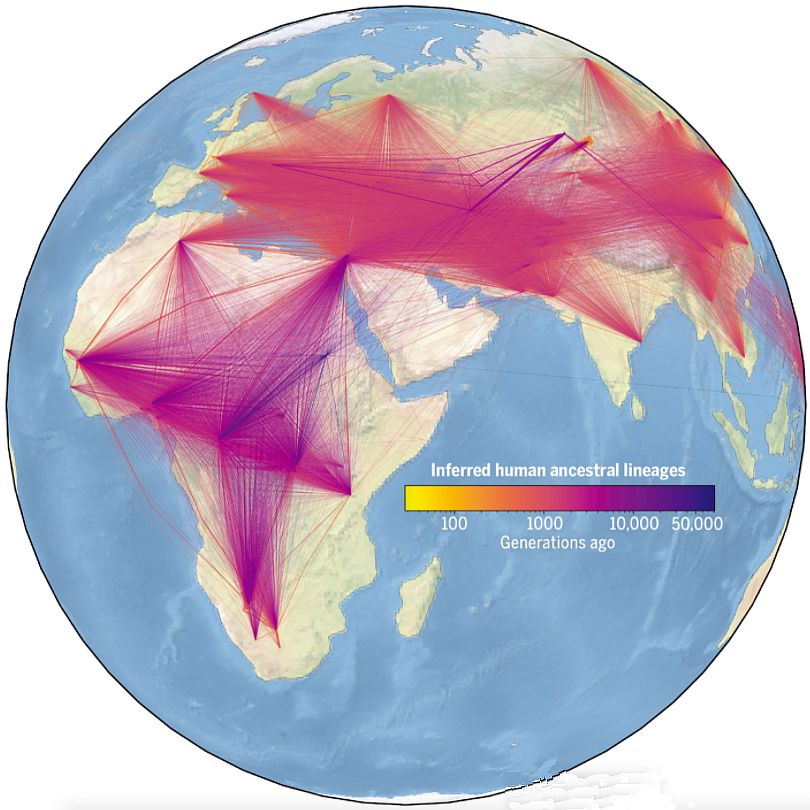
Thank you for writing in with this question. This is a subject I have been interested in for decades. I’ve only been waiting for enough genetic data to accumulate so that we could build a family tree of all people, going right back to Adam and Eve. Sadly, this paper, and the popular-level reporting on it, claim that the ancestry of modern humans is in Africa. The images associated with the paper led many people to think this supports the Out of Africa model of human origins (figure 1). Yet, we can answer these challenges while at the same time building trust in the Bible.
It is also a brand new paper, so it should not be surprising that the answer was not on creation.com, yet!
However, your simple question involved hours of research on my part. I read the article you supplied,1 then a summary article in Science by Rees and Andrés,2 then the full article in Science by Wohns et al.,3 then the publicly accessible supplementary materials (69 pages before the 208 references).4 I then had to spend hours writing and thinking. I wanted to do this, mind you, but I was not sure where I was going to find the time. By you sending this request for information to CMI, I had all the justification I needed to dig deeply into one of my favourite subjects: genealogy and genetics. For that, I thank you.
What they did
Wohns et al. started with a data set of thousands of modern human genomes. They combined the results of the 1000 Genomes Project, the Human Genome Diversity Project, and the Simons Genome Diversity Project. This was not easy to do, for each database covered slightly different portions of the human genome and the data are of variable quality, with the 1000 Genomes Project being much lower quality than the others. Data quality is a real problem for geneticists. About a decade ago I was studying a high-coverage genomic dataset of a single family that included four grandparents, two parents, and eleven children. This was supposedly the ‘platinum standard’ of gene sequencing. Yet, I found the data replete with errors. I was trying to measure gene conversion, where single letters are copied from one parental chromosome to the other before being passed to the child, but there were too many conflicting places where a child popped up with a variant in one of the grandparents but in neither parent or where a guaranteed variant did not appear in a child. This was frustrating. I had to table the project because the error rate was simply too high. I could easily map all the chromosomal recombinations across the three generations, but not gene conversion. I appreciate the work this team did to overcome such limitations.
They also included high-quality (at least, the highest quality available) DNA from several ancient DNA specimens. This included four ‘modern’ individuals (two parents and their children) that were buried in southern Siberia about 4.6 KYA,5 three Neanderthals (ranging from 50 to 80 KYA6,7,8), and one Denisovan (from about 80 KYA9).10 An additional 3,592 ancient DNA samples of lesser quality were used to help ‘date’ the first appearance of different genetic variants. They did not specify the range of dates for these individuals, and I did not want to look up the information in the dozens of papers from which the data were drawn, but many titles include the phrases “Bronze Age”, “Neolithic”, and “Ice Age”.11 Thus, we are not talking about genomes from even 100 KYA. This will become important later.
Once the data were in hand, they created a family tree for each of the over six million variable sites in the genome. After this, they combined these trees into an average framework. The result is what they called a “family tree” but note that this is not a list of names. It is a genetic approximation of ancestry only. They had to infer a lot of the information, especially when more than one possible tree could explain the same data, and this happened often.
They also focused their attention on chromosome 20 only. This was done for the simple fact that this is one of the smallest chromosomes. My colleagues and I took a similar approach in a paper on the distribution of alleles in the modern human population.12 Yet, even when limiting the data to chromosome 20, they still had to calculate the time to the most recent common ancestor (TMRCA) for about 300 billion pairwise combinations of alleles.
What they claimed
The main claim is that, by taking thousands of human genomes (both modern and ancient), one can build a worldwide family tree. This is true, sort of. The implications are that humans started in Africa tens of thousands of years ago. This is not true, but the assumptions they made before they began their study drove them to these conclusions.
They also found evidence for recent population bottlenecks in most of the sampled populations. In other words, the family trees of the people in each area collapsed into a very small group of ancestors. This could be due to lots of local inbreeding, which is expected, given human behaviour. Alternatively, it could be due to these populations being founded in more recent times by a smaller number of people than they assumed.
They also detected many recurrent mutations and “pervasive signals of sequencing error.” These were not strong enough to affect their results and they took steps to clean up the data before running any analysis.
The signal for Denisovan ancestry among Papuans (the indigenous people of the large island of New Guinea) was also very strong. This is interesting in itself, but please note that the earlier an event happened in human history, the less likely anyone will be able to accurately describe it. You cannot tell when this interbreeding event happened, because carbon dating is highly problematic that close to the Flood. You also cannot tell where it happened, because the people were actively migrating across the world from Babel. At some point in those early generations, people from two very dissimilar family lines met and married. That’s all we know, but the evidence is clear that it did happen. This raises huge questions about the Out of Africa event. We were supposed to have evolved from Homo erectus into Homo sapiens in Africa. Neanderthals and Denisovans were not part of this. Yet, when we supposedly left Africa, we met and successfully mated with these non-humans? Consider that 10% of the genomes of some of the people in Oceania is “non-modern” DNA,13 yet they are fully functioning, highly intelligent, and creative human beings. You would never have known that a large fraction of their ancestry traces back to non-humans. Humans arose in Africa? Baloney!
Problems with comparing genealogy and geography
You can’t necessarily correlate geography, genealogy, and genetics. This is a long-standing and well-known problem. First, people can migrate long distances over their lifetimes. Thus, just because you find a person buried somewhere does not mean they were born there. Second, just because you find an ancient person that carries a genetic variant does not mean that this was the first person to have it. It only means that this is the first person known to carry it. You can’t truthfully claim that this person is the ancestor of anyone alive today. Third, these speculations are derived from computer models, which are necessarily abstract. You cannot necessarily draw firm conclusions after your computer pops out with some number.
Recently, a different group of scientists tried to claim that they had identified the location where modern humans first arose. They discovered some deep branches in the human mitochondrial family tree and realized that everyone known to carry these mitochondria lived south of the Zambezi River in southern Africa.14 They went so far as to claim humans emerged in the paleo-Okavango wetland (the modern Okavango is famous for being a ‘river to nowhere’ and has been featured in multiple nature documentaries). These conclusions have multiple problems. First, even if the mitochondrial ancestor could be traced to this location, this does not mean that the Y chromosome ancestor or the many ancestors that contributed to the autosomes would have lived in the same place. Second, just because people with these mitochondria live in a certain place today does not mean their ancestors lived in this same place in the ancient past.15 This is especially true given the evidence for a recent expansion of the Bantu-speaking people across eastern and southern Africa,16 with the extirpation of most of the original inhabitants of the current Bantu lands.17 Third, the entire argument relies on evolutionary assumptions about deep time and molecular clocks. There is no reason to accept these claims.
Wohns et al. are also claiming the ability to place ancestors on a map, but this is solely based on where the people are found today. They are also placing the ancestral root population of modern humanity far up in the northeast corner of Africa. This is quite far from where most evolutionists would place our homeland (see above). It is also uncomfortably close (for them) to the Middle East, where the Bible claims we originated. To be fair, they used the DNA of ancient people to ‘ground truth’ the data, but there are still huge assumptions at play. They are also aware of the limitations of this approach, saying:
“ … the accuracy of any ancestral geographic inference method will be limited when the distribution of sampled individuals does not reflect the location of the samples ancestors.”
and
“ … migrations occurring within the past few thousand years [references] means that present-day distributions of groups in Africa and elsewhere may not represent those of their ancestors, and thus we may have a distorted picture of ancient geographic distributions.”
Yet, a mass migration, where the death location of many individuals is incongruent with their birth locations, is exactly what the biblical account of the Tower of Babel would predict. Thus, if Babel occurred, their results are necessarily spurious.
They also understand that there is some uncertainly in their predicted timing of events, saying, “We caution that the absolute ages we report have some degree of error,” but they also claim their estimates should only be up to 16% off, and mostly with an upward bias. Yet, their timing of events is based squarely on the accuracy of carbon dating, which they admit to in the supplementary material. Creationists have long said that carbon dates become magnified the closer one gets to the Flood. The main reason for this is that the earth’s magnetic field is decreasing exponentially over time. Since the production of carbon-14 in the upper atmosphere is caused by cosmic rays, and since the strength of the magnetosphere controls how many cosmic rays can get into the atmosphere, extrapolating this backward tells us there was less carbon-14 in the distant past. Thus, any material from the early post-Flood years will have an anomalously high carbon date because it will have unusually low levels of carbon-14.



Also, you can see the effects of their geographic assumptions in the data they presented. For example, in a publicly accessible video,18 they show how the connectedness of all populations grows over time. In this screenshot (figure 2), from near the end of the simulation (which runs backwards in time), notice how the plotted points run straight across the Red Sea and Northern Arabia. This is extremely artificial, as any migration route would be expected to go around the north of the Red Sea (via Sinai or the Mediterranean coast) or across the 10-mile-wide Bab al-Mandab strait between Djibouti and Yemen. This distribution of points is an artefact of their method of plotting ancestral locations as the midpoint of a great circle route between two daughter nodes. While interesting from an academic point of view, this is entirely non-historical.
There are other problems that the presentation of the data reveals. For example, there appear to be genetic links streaming from Oceania toward Africa hundreds of thousands of years ago (in figure 2, the clock shows a date of 837.5 KYA). This is long before the Out of Africa Event (which they date to ~70 KYA) and much farther back in time than any of their genomic samples. What we are seeing is simply the results of a simplistic computer model. It has something to do with Denisovan ancestry, but it has no basis in reality.
In the mid-point of the simulation (figure 3), the supposed migration path out of Africa is more well defined. This is entirely due to the placement in time and geography of the modern and ancient samples incorporated into the study. If they did not have samples from the area around Israel, there would not be a nexus at that location at these later times.
The initial populations are visible at the beginning of the model run (figure 4). It was from these, guided by the alleles seen in ancient genomes, that they claim to have traced all people back to a single source.
Their methods also create some contradictions. For example, they claim people arrived in Papua New Guinea 100,000 years before the earliest evidence for humans in the region (I believe their video indicates a much earlier timeframe). They also date the arrival of people in the Americas long before the current consensus date. This could indicate problems in archaeology, or it could be telling us something is wrong with their methods.
Generation times
I may have missed it, but I was not able to determine how they defined one “generation”. This has an obvious definition in the real world, but the definition in the world of computer simulations is not set in stone. They may have assumed an average generation time. Most evolutionary studies have used “20 years” as the average, but the real value is closer to 30.19 Yes, human females can have children when they are young, but they can also have children up to the age of about 40. The average is thus much greater than the minimal value.
Alternatively, they may have used some estimate of recombination rates as a proxy for generation time. They were building millions of trees, including trees for letters that are near to each other in the genome. Given one to two crossovers per long chromosome arm per generation (the rate varies for each chromosome arm), and assuming crossover locations are random (which is not true), one can estimate how long it would take, on average, to split two letters, given how far apart they are along the chromosome arm. Their conclusions also depend on an identical recombination rate across all populations, yet some data suggest that recombination rates are higher in Africans than other populations.20 That alone throws a giant monkey wrench into their study. I also wonder how the newly emerging consensus that there was deep population subdivision in Africa in ancient times might affect their conclusions.21
They ignored the lifespans of the biblical Patriarchs (and unnamed matriarchs), the biblical claim that the entire human population came from three founding couples, that the source population was not in Africa, and the potential effects of Patriarchal Drive. Applying the biblical model to these same data would generate entirely different conclusions.
In the end, this is just a fancy new application of the same old arguments. We already knew that there was more diversity in Africa, that recombination blocks were smaller in Africa, and that the “deepest-rooting” Y and mitochondrial lineages existed in Africa. Yet, they made several sweeping assumptions that are driving their main conclusions but which we may openly question. This is not ‘proof’ that we came out of Africa.
References and notes
- Guy, J., DNA reveals biggest-ever human family tree, dating back 100,000 years, msn.com/en-us/news/technology/dna-reveals-biggest-ever-human-family-tree-dating-back-100000-years/ar-AAUgyub, 17 Feb 2022. Return to text.
- Rees, J. and Andrés, A., Inferring human evolutionary history, Science 375(6583):817–818, 2022. Return to text.
- Wohns, A.W. et al., A unified genealogy of modern and ancient genomes, Science 375(6583):eabi8264, 2022. Return to text.
- science.org/doi/10.1126/science.abi8264. Return to text.
- Sequencing details are given in Table S1 of Wohns et al. Return to text.
- Prüfer, K. et al., The complete genome sequence of a Neanderthal from the Altai Mountains, Nature 505(7481):43–49, 2014. Return to text.
- Prüfer, K. et al., A high-coverage Neandertal genome from Vindija Cave in Croatia, Science 358(6363):655–658, 2017. Return to text.
- Mafessoni, F., et al., A high-coverage Neandertal genome from Chagyrskaya Cave, PNAS 117(26):15132–15136, 2020. Return to text.
- Meyer, M. et al., A high coverage genome sequence from an archaic Denisovan individual, Science 338(6104):222–226, 2012. Return to text.
- These are, of course, the evolutionary dates. KYA = thousands of years ago. Return to text.
- See reichdata.hms.harvard.edu/pub/datasets/amh_repo/curated_releases/index_v42.4.html. Return to text.
- Sanford, J. et al., Adam and Eve, designed diversity, and allele frequencies; 8th ICC, pp. 200–216, 2018; digitalcommons.cedarville.edu/icc_proceedings/vol8/iss1/8/. Return to text.
- Larena, M. et al., Philippine Ayta possess the highest level of Denisovan ancestry in the world, Current Biology 31:1–12, 2021. Return to text.
- Chan, E.K.F. et al., Human origins in a southern African palaeo-wetland and first migrations, Nature 575(7781):185–189, 2019. Return to text.
- Bergström A. et al., Origins of modern human ancestry, Nature 590(7845):229, 2021. Return to text.
- Tishkoff, S.A. et al., The genetic structure and history of Africans and African Americans, Science 324(5930):1035–1044, 2009. Return to text.
- Prendergast, M.E. et al., Ancient DNA reveals a multi-step spread of the first herders into sub-Saharan Africa, Science 365(6448):eaaw6275, 2019. Return to text.
- Available in their supplementary data. See reference 4. Return to text.
- Langergraber, K.E. et al., Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution, PNAS 109(39):15716–15721, 2012. Return to text.
- Hinch, A.G. et al., The landscape of recombination in African Americans, Nature 476(7359):170–175, 2011. Return to text.
- Lipson, M. et al., Ancient DNA and deep population structure in sub-Saharan African foragers, Nature 603(7900):290–296, 2022.Return to text.






Readers’ comments
Comments are automatically closed 14 days after publication.