The four dimensional human genome defies naturalistic explanations

The human genome is the most complex computer operating system anywhere in the known universe. It controls a super-complex biochemistry that acts with single-molecule precision. It controls the interaction network of hundreds of thousands of proteins. It is a wonderful testament to the creative brilliance of God and an excellent example of the scientific bankruptcy of neo-Darwinian theory. Why? Because the more complex life is, the less tenable evolutionary theory becomes. Super-complex machines cannot be tinkered with haphazardly or they will break. And super-complex machines do not arise from random changes.
I am serious when I compare the genome to a computer operating system. The only problem with this analogy is that we have no computers that can compare to the genome in terms of complexity or efficiency. It is only on the most base level that the analogy works, but that is what makes the comparison so powerful. After millions of hours of writing and debugging we have only managed to create operating systems that can run a laptop or a server, and they crash, a lot. The genome, though, runs a hyper-complex machine called the human body. The organization of the two are radically different as well. A team made up of computer scientists, biophysicists, and experts in bioinformatics (in other words, really smart people) compared the genome of the lowly E. coli bacterium to the Linux operating system (figure 1) and have discovered that our man-made operating systems are much less efficient because they are much more “top heavy”.1 It turns out that the bacterial genome has a few high-level instructions that control a few middle-level processes, that in turn control a massive number of protein-coding genes. Linux is the opposite. It is much more top heavy and thus much less efficient at getting things done. The bacterium can do a lot more with fewer controls. I predict that the study of genomics will influence the future development of computers.
Also, our computers use comparatively simple programs. Programmers talk about “lines of code”. We all learned in math class that a line is a one-dimensional object. So our computer programs are essentially one-dimensional. The human genome operates in four dimensions. This is one of the greatest testimonies to the creative brilliance of God available.

The First Dimension: the DNA molecule
The human genome is about 1.8m long. All of it fits into the cell nucleus. To put that in perspective, if you were to make your DNA as thick as a human hair, you would then have more than 50 kilometres of DNA, crunched up into something about the size of a golf ball. Already, we must understand that God is an incredible engineer.
If we were to look at the sequence of letters in the DNA, it might look like this:
That is the first 700 letters of the human Y chromosome. Not very impressive, is it? But if we take that same sequence and replace the four letters with four coloured pixels, you get something that looks like figure 2.
The first dimension of the genome is simply the order of the letters. They spell out genes and those genes tell the cell to do things. This is not really that complicated, but things are about to change.
The Second Dimension: the interaction network
The second dimension of the genome deals with the way one section of DNA interacts with another section. As we have already seen, you can draw the first dimension out easily enough. But if you tried to draw out the second dimension you would first need to draw many arrows connecting different parts of the linear string of DNA. It would be impossible to draw the entire interaction network of the genome, so a small example will have to suffice. Micro RNAs (miRNA) are very small molecules (about 22 nucleotides) that are involved in the regulation of gene function. Figure 3 shows a portion of the miRNA regulation network as it acts on just 13 genes that are upregulated in association with atherosclerosis (hardening of the arteries). These genes are targeted by 262 miRNAs, creating 372 “regulatory relationships”. Not included in figure 3 are the 33 other genes that are downregulated by 295 miRNAs when the body is dealing with this condition. Remember, this is only a small slice of the 2nd dimension of the genome!
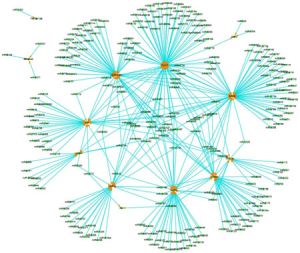
The second dimension deals with things like specificity factors, enhancers, repressors, activators, and transcription factors. These are proteins that are coded in the DNA, but they move to another part of the genome after they are made and turn something on or off. But there are additional things happening in this dimension. During the process of protein manufacturing, a gene is “read” by the cell during a process called transcription. Here, the DNA is copied into a molecule called RNA. The RNA is then translated into a protein. We have an excellent animation of this process on our multimedia site. But in a process called post-transcriptional regulation, the RNA can be inactivated or activated by other factors (like miRNAs) coded elsewhere in the genome.
The massive, multi-million-dollar ENCODE project revealed something about the genome that we are still trying to fully grasp. One of the greatest mysteries is how only about 22,000 genes can produce more than 300,000 distinct proteins. The answer is that the cell goes through a process called alternate splicing, where the genes are sliced and diced and different parts are used by different cells at different times and under different circumstances to produce the many different proteins. This incredibly complex process is just one part of that second dimension of the genome.
The Third Dimension: 3-D DNA architecture
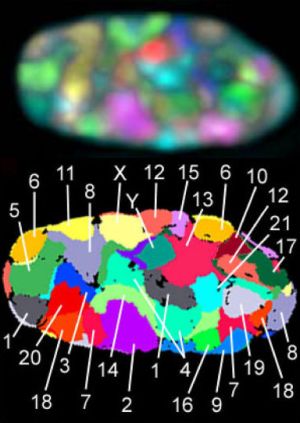
The third dimension deals with how the shape of the DNA molecule affects the expression and control of different genes. We have learned that sections of DNA that are buried deep within the coiled-up DNA cannot be activated easily.4 So genes that are used often are generally easily accessible. Thus, when God wrote out the information in the genome along that one-dimensional strand, He intentionally put things in a certain order so that they would be in the correct place when the DNA was folded into a 3-D shape.
One of the big revelations of the Human Genome Project was that genes that are used together do not necessarily appear near one another in the genome. Claims like “It’s just junk” and “The genome is nothing more than millions of years of genetic accidents” were raised. This did not last long, however, for once people started looking into how the genome is organized in the nucleus,5 they realized that, not only does each chromosome have a specified position in the nucleus, but genes that are used together are generally found next to each other in 3D space, even when they are found on different chromosomes!
The Fourth Dimension: changes to the first three dimensions
The fourth dimension of the genome deals with the way the first three dimensions change of the fourth dimension, time. Yes, you read that right. The shape (3rd dimension), interaction network (2nd dimension), and the sequence of letters (1st dimension) all change. This so far outstrips even our most modern computers that the analogy isn’t even fair any more.
This fourth dimension can be illustrated in several ways. We know that different liver cells have different chromosome counts.6 This is due to the fact that the liver needs lots of copies of certain genes that are involved in metabolism and detoxification. Instead of filling the genome with many copies of these genes, the liver just makes copies of them for its own use. We also know that different brain cells have different number and locations of various transposons.7 These are the “jumping genes” that are thought by evolutionists to be leftovers from ancient viral infections. Problem is, they are vital for the development of the human brain. Did you catch that? The genome dynamically reprograms itself. This is something that computer scientists have long struggled with. How can you make a self-modifying code that does not run out of control? We also know that transposons are critical for controlling embryonic development in the mouse.8 So much for calling them “junk DNA”!
Conclusions
The genome is a multi-dimensional operating system for an ultra-complex biological computer, with built-in error correcting and self-modification codes. There are multiple overlapping DNA codes, RNA codes, and structural codes. There are DNA genes and RNA genes. The genome was designed with a large amount of redundancy, on purpose, by a highly-intelligent being who used sound engineering principles during its construction. Despite the redundancy, it displays an amazing degree of compactness as a mere 22,000 or so protein-coding genes combinatorially create several hundred thousand distinct proteins.
I have a challenge for the evolutionist: Explain the origin of the genome! Charles Darwin wrote in the origin of species:
If it could be demonstrated that any complex organ existed, which could not possibly have been formed by numerous, successive, slight modifications, my theory would absolutely break down.
I know this quote has been abused (by both sides of the debate) but let’s think about this for a second. The simpler life is, the easier it is to explain in Darwinian terms. On the other hand, the more complex life becomes, the more intractable a problem it causes for evolutionary theory. We have just seen that the genome is the opposite of simple. This should make all Darwinists very uncomfortable.
I claim the genome could not have arisen through known naturalistic processes. The evolutionist who wants to take up this challenge must give us a workable scenario, including the source of informational changes, an account of the amount of mutation necessary, and a description of the selective forces necessary, all within the proper time frame. They will discover that evolution cannot do what they require, even over millions of years.
By the way, this was a brief summary of the information contained in the DVD The High Tech Cell. If you want more information, I suggest you purchase a copy from our webstore.
References and notes
- Yan, K.-K., et al., Comparing genomes to computer operating systems in terms of the topology and evolution of their regulatory control networks. PNAS 107(20):9186-9191, 2010. Return to text.
- Seaman, J., and Sanford, J., Skittle: a 2-dimensional genome visualization tool. BMC Bioinformatics 10:452, 2009. Return to text.
- Lin, M., Zhao, W., and Weng, J., Dissecting the mechanism of carotid atherosclerosis from the perspective of regulation, International Journal of Molecular Medicine 34:1458-1466, 2014. Return to text.
- van Berkum, N.L., Hi-C: a method to study the three-dimensional architecture of genomes, Journal of Visualized Experiments 6(39):1869, 2010. Return to text.
- Bolzer, A., et al., Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes, PLoS Biol 3(5):e157, 2005. Return to text.
- Duncan, A.W., et al., The ploidy conveyor of mature hepatocytes as a source of genetic variation, Nature 467:707-710, 2010. Return to text.
- Baillie, J.K., et al., Somatic retrotransposition alters the genetic landscape of the human brain, Nature 479:534-537, 2011. Return to text.
- Tomkins, J., 2012. Transposable Elements Key in Embryo Development; icr.org/article/6928, July 25, 2012. Return to text.





Readers’ comments
Comments are automatically closed 14 days after publication.