Journal of Creation 25(2):118–124, August 2011
Browse our latest digital issue Subscribe
The family of cats—delineation of the feline basic type
Based on previously reported hybridizations, cats have long been considered to belong to a single basic type.1 However, there has been discussion concerning whether great cats and small cats might represent independent sister clades. Recent DNA sequencing data confirm that such distinctions are not fundamental in nature and that all cats share a common genetic ancestry. More recently described hybridizations between great cats and small cats, along with various other studies described in the present article, further support the hypothesis that all cats belong to a single clearly delineated basic type. The Nimravidae (paleosabers), the Machairodontinae (neosabers) and the genus Panthera, each underwent a prominent radiation during the tertiary period. All three taxa represent cat-like placental carnivores, and they may all have arisen from the same basic type.
Cats and their taxonomic position

The cat family is placed within the order Carnivora, which comprises nine extant families (or ten if mongooses are considered a separate family; Herpestidae). The carnivores are grouped into two suborders: the catlike carnivores, or Feliformia, including the Felidae (cats), the Hyaenidae (hyenas), the Viveridae (civets), and the Herpestidae (mongooses); and the dog-like carnivores, or Caniformia, including the Canidae (dogs), the Ursidae (bears), the Procyonidae (raccoons), and the Mustelidae (weasels), as well as two marine families, the Otariidae (sea lions) and the Phocidae (seals). The present role of carnivores in nature is regulatory, keeping in check the numbers of herbivores. They are assumed to indirectly help maintain healthy populations of herbivores by selectively devouring non-healthy and phenotypically disadvantaged animals. The carnivores share a relatively homogeneous phenotype. Many are capable of running quickly, possess conspicuous canine teeth often used for catching and killing prey, and display the carnivore-typical carnassial teeth, which include the last premolars of the upper jaw and the first molars of the lower jaw. Instead of having a grinding surface, these teeth have a flattened, razor-like crown used for slicing through muscle tissue when devouring prey. In the omnivorous carnivores, such as bears, true carnassial teeth do not develop.
The 38 species of extant cat have a very characteristic phenotype readily distinguished from other species of animals, even by laymen. Recently, the clouded leopard (Neofelis nebulosa) was separated into two species, which, if acknowledged, brings the total number of species to 39.2 They possess a lithe, muscular, compact and deep-chested body. Technical diagnostics include: pointed, elongate canine teeth; large carnassials, strongly shearing in function; the dental formula 3/3, 1/1, 2-3/2, 1/1; ossified auditory bullae, inflated in appearance and divided by a bilaminate septum (except Leopardus jacobita, the Andean mountain cat, which has a double-chambered bulla);3 a tongue covered with numerous, horny papillae that are directed backwards; digitigrade extremities with five toes on the forefoot and four on the hindfoot; claws that are sharp, strongly curved, and usually highly retractile, protected by a fleshy sheath (except in the genus Acinonyx, the cheetahs).4
One or two family histories?
The earliest cat-like carnivore family, the Nimravidae (paleosabers), includes two lineages that suddenly appear in the late Eocene jungles of North America. One lineage, represented by Hoplophoneus, was saber-toothed; the other lineage, represented by Dinictis, was rather like a modern serval. So convincingly cat-like are these fossils that originally they were called paleofelids and designated the first cats.4 The more recent cat family, the Felidae, include the modern cats, Felinae; and the neosabers, Machairodontinae, e.g. Smilodon.5 The Nimravidae and the Felidae display only modest skeletal differences. Nevertheless, they are placed into two distinct families. The most prominent skeletal difference is that nimravid fossils lack a bony wall (septum) in their middle-ear chambers (auditory bullae) or sometimes the whole chamber, implying these structures were cartilaginous.6 The nimravid, Barbourofelis, which lived contemporaneously with the Felidae, had ossified auditory bullae, but there is no evidence of a bony septum.7 Such morphogenetic differences require modest selective change. Indeed even in extant cats the auditory bullae are first cartilaginous and only later ossify. As versatile as ossification of the auditory bullae has proven to be in helping define extant cat species, calling an extinct animal a non-cat that is otherwise clearly a cat, simply for want of evidence of such ossification, seems excessive.
The Nimravidae-Felidae ‘two-family’ hypothesis is supported less by ossifications and more by phylogenetic considerations; specifically, that the Felidae arose from Proailurus during the Miocene in the Old World.8 Proailurus, the animal currently nominated the first ‘true’ cat, was short, only 15 cm in length. It had very many civet-like features and was probably plantigrade (walking on the soles of its feet) and not digitigrade (walking on its toes) like cats today.8 Perhaps the need for a link to a postulated ‘common-civet ancestor’ is the primary motivation for trying to classify Proailurus among the cats. It had been previously classified among the civets (Viverridae) and there is much to support this classification still; plantigrade gait, flatter skull, extra teeth, the bones of Proailurus are very similar to the living viverrid, Cryptoprocta, the fossa of Madagascar.2,5,7 The next most recent cat, Pseudaelurus, had a clearly cat-like skull, based on its general morphology and dentition. It roamed Europe and North American during the Miocene. Several different forms of Pseudaelurus existed and these are believed to have formed the basis for the later radiation and diversification of the neofelid cats and the sabre-tooth tigers.9 Proailurus, from the Miocene, is the reason why New World paleosabers from the Eocene cannot be cats. As a result the origins of the paleosabers have had to be placed elsewhere; e.g. within the dog-like carnivores9 or even outside the carnivores10. It is easy to see how the two-family hypothesis could be specious, based on preconceived notions.
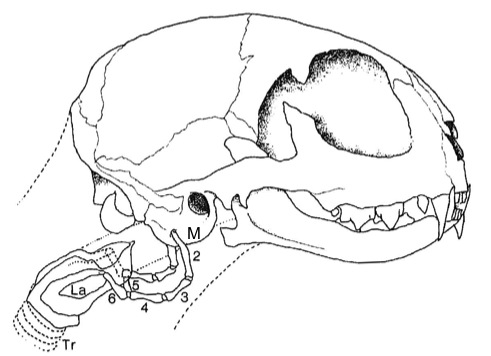
Differences in ossification of skeletal features are not necessarily diagnostic of family status. A case in point is the early attempt to separate small cats and big cats into separate subfamilies based on ossifications in their hyoid apparatus and their ability to roar.7 Sir Richard Owen11 was the first to explore hyoid anatomy and felid vocalization. The hyoid apparatus consists of two chains of seven bones, joined at the fifth (basihyoid). These stretch from the auditory bullae to the larynx, support the voice box, and confer characteristic vocalizations (see figure 1). In small cats (the felines) the third bones (epihyoids) are ossified and short, and they cannot roar. In the larger big cats (the pantherines) the third epihyoids are cartilaginous and extended, and all but one of these cats can roar. In snow leopards (P. uncia; figure 2) the epihyoids are cartilaginous; however, they still cannot roar because they lack additional pads in their vocal folds.12,13 Ossification of the seven hyoid bones in felids is never complete. Both the first (tympanohyoid) and seventh (chondrohyoid) ‘bones’ are cartilaginous. These attach to the auditory bullae and cartilaginous larynx, respectively.13 Bone ossifications are of less taxonomic importance than genetic and hybridization data, which support the conclusion that small cats and big cats belong in the same subfamily.
It is submitted that lack of ossification of the auditory bullae in the Nimravidae is similarly of questionable diagnostic use for the family’s phylogenetic position and that the Nimravidae represent at least a sister clade of the Felidae, if not actual members of the same basic type. Their distributions reflected the dominant biomes. During the course of the Tertiary period, gradual cooling and drying caused major shifts in these biomes. During the Eocene/Oligocene transition, subtropical rainforests were replaced by woodland savannas.14 These more open forest conditions resulted in adaptive radiations in herbivore groups including oreodonts, anchitheres and small rhinocerotoids (Janis 1998). Since these were the principle prey of the Nimravidae, adaptive radiation in the herbivores would account for the adaptive radiation observed in the paleosabers during that period. Similarly, at the Miocene/ Pliocene transition, grassland savannas were replaced by prairies.15 Adaptive radiations of cursorial herbivore groups, including camelids, antilocaprines, and equines was then accompanied by adaptive radiation of their predators, the Felidae.15 Today both the paleosabers (Nimravidae) and neosabers (Machairodontinae) are completely extinct. Only in the extant clouded leopard (Neofelis nebulosa) can a vestige of the saber tooth phenotype still be observed; although both the upper and the lower canines are pronounced, this is a unique occurrence in the cat family. Why these animals became extinct is something of a mystery. These large carnivores would have required large prey; saber teeth are inefficient for capturing and killing small prey. Biome change may have led to an imbalance in predator–prey relations, resulting both in the loss of sustainable populations of large prey and in the extinction of these remarkable cats.
Pelage pattern—a common origin
The development of melanistic coat colour patterns within the cat family is not entirely understood, but some interesting conclusions can be drawn. Weigel16 believed that all of the extant melanistic coat colors and patterns arose from a single original type of relatively large dark spot. Werdelin and Olsson17 replaced Weigel’s proposal with evidence indicating that the original cat family coat pattern was actually small spots or ‘flecks’. This is highlighted by changes observed in pelage patterns of jaguars and leopards during their development. Juveniles possess a simple spot pattern but adults display a range of complex rosettes. A recent theoretical study by Liu and colleagues,18 using mathematical models, confirmed that in both the jaguar and leopard a single mathematical function (based on a Turing-Model) could simulate both the simple fleck-pattern of juveniles and the complex rosette-pattern of the adults. It appears that differences in pelage patterns can be traced back to subtle variations of the basic pattern. These can adequately explain the changes observed during both the development of individuals and the radiation of cat lineages. In other words, mechanisms of change such as recombination and natural selection appear sufficient to explain the variation in melanistic spot patterns observed within the cat family. Other patterns like the stripes of the tiger were not investigated in the study, but the same kind of mathematical modeling has been used to simulate formation of stripes in other animals. Whole coat melanism in the domestic cat, jaguar and jaguarundi has been elucidated at the molecular level.19 These pelage pattern studies seem to reinforce the general observation that cats form a unique and well-defined group of animals.
Radiation of modern cats in the Miocene
The phylogenetic history of extant cats is complex and has been controversial for a long time, primarily because of the extremely rapid speciation events that took place during the Tertiary period. Recently, based on DNA sequencing studies, relations between the various lineages and species could be clarified. DNA analyses of a spectrum of genes, along with modern statistical techniques, have revealed that the extant cat family can be subdivided into eight lineages (figure 3a):

- The Panthera Lineage (lion, jaguar, leopard, tiger, snow leopard, and cloudy leopard)
- The Bay Cat Lineage (bay cat, Asian golden cat, and marbled cat)
- The Caracal line (caracal, African golden cat, serval)
- The Ocelot Lineage (ocelot, margay, Andean mountain cat, pampas cat, Geoffroy’s cat, kodkod, and tigrina)
- The Lynx Lineage (Iberian lynx, Eurasian lynx, Canadian lynx, and bobcat)
- The Puma line (puma, jaguarundi, and cheetah)
- The Leopard Cat Lineage (Pallas’ cat, rusty spotted cat, Asian leopard cat, fishing cat, and flat-headed cat)
- The Domestic Cat Lineage (domestic cat, European wildcat, African wildcat, Chinese desert cat, desert cat, black-footed cat, and jungle cat).

In spite of the excellent general consensus, a few phylogenetic relations are still uncertain. The placing of the Andean mountain cat (Leopardus jacobita) within the Ocelot Lineage, the placing of the jungle cat (Felis chaus) and the black-footed cat (F. nigripes) within the Domestic Cat Lineage, and the hierarchy of the Panthera Lineage are all questioned. Presence of a large non-functional mtDNA translocation in the nuclear genome of all extant members of Panthera supports a late Pliocene radiation of the genus,20 and a recent cladistic study has thrown much light onto the detailed relationships between extant and extinct Pantherinae.21 In the cat phylogeny of Johnson et al.,22 21 of 36 speciation branch points took less than 1 million years each, and seven speciation events that usher in the eight major cat lineages took on average just 600,000 years to complete. Brief and spectacular radiation events appear to be the norm in most vertebrate phylogenies. It is the reason why large numbers of genes must be employed in phylogenetic studies. During the radiation of the cat family, significant migrations took place across continents and distribution zones. These are displayed in figure 3b.23
The taming of the cat
Felis catus, the domestic cat, is believed to have arisen from the African wildcat, F. lybica, and not from the European wildcat, F. silvetris. Although taxonomists sometimes lump these two wild species together, a number of phenotypic differences are evident. F. lybica is easier to tame; it is far less shy of man. In appearance, F. lybica is somewhat larger and stockier than F. silvestris. Felis lybica also has a black mark on the sole of its feet, continuing between the toes. The domestic cat also displays a number of differences from F. lybica, its ancestral species. Domestic cats tend to be smaller. They have a smaller brain and longer digestive tract. Domestic cats are also tamer than their wildcat cousins. At least two features contribute to this. First, their appearance, including their behavior, is more akin to the juvenile animal. Second, and perhaps related to this, the relative size of their adrenal glands is smaller. The adrenal glands produce epinephrine (adrenaline), and when animals are stressed this causes the fight-or-flight behavior so familiar in wild animals. Because domestic cats have a smaller adrenal gland, less epinephrine is produced and the animals are much quieter than wild cats.23 These studies parallel observations by Trut et al.24 in studies of fox domestication. Together the studies suggest that the domesticated phenotype lies dormant within the genetic potential of many wild species. A recent extensive study of mitochondrial DNA sequences from hundreds of domestic and wild cats across Europe, Asia, and Africa (including the domestic cat (F. catus), the European wildcat (F. silvestris), the African wildcat (F. libyca) and the Chinese desert cat (F. bieti), using the desert (sand) cat (F. margarita) as an outgroup, has clarified with great certainty the various ancestral relationships. The domestic cat is clearly descended from the African wildcat (F. libyca or F. silvestris libyca), not the European wildcat (F. silvestris or F. s. silvestris). In addition, the domestic cat, the European wildcat, the African wildcat, and also the Chinese desert cat all appear to belong to a single species. Cementing this idea is the fact that mitochondrial DNA from domestic cats is routinely found in all three wild cat populations. Natural hybridizations between them are frequent events.25
Hybridizations within the cat family

Cats are beloved zoo and house animals, so there are many reports of hybridizations either occurring spontaneously or deliberately undertaken (figure 4),26 Seven of the eight major cat lineages reported by Johnson et al.23 are linked by hybridizations. Only the Bay Cat Lineage has not been linked by hybridization to another lineage. Phenotypically, however, it is closely related to the Caracal lineage. Both the lineages contain golden cats. The Bay Cat Lineage includes the Asian golden cat, and the Caracal Lineage includes the African golden cat. Moreover, in the Johnson et al. study they are placed as sister groups in the phylogenetic DNA-sequence tree. All fourteen interlineage hybrids can be traced to one of three critical species: the domestic cat (F. catus), the bobcat (L.rufus), and the puma (P. concolor). Besides interlineage hybrids, numerous intralineage hybrids have also been reported. Alderton4 describes a cross between a lion and the hybrid of a jaguar and a leopard. The bobcat hybridizes with all the lynxes (see figure 4). There is clear evidence that hybridizations occur between the major cat lineages and that hybridizations occur within the lineages; therefore, the hypothesis—all extant felids belong to a single basic type—has been reasonably proven.
Hybrids between cats are often given unique names. Many of these names are contracted fusions of their common names; the first part indicating the male parent and the second part, the female parent. These include: P. leo × P. tigris (Liger or Tigon), P. leo × P. pardus (Liard or Leopon), P. tigris × P. pardus (Tigard), P. onca × P. pardus (Jagulep or Lepjag), P. onca × P. leo (Jaglion), P. concolor × P. pardus (Pumapard), C. serval × C. caracal (servical or caraval), L. wiedii × L. pardalis (marlot). Female hybrids of lions and tigers are fertile and reproduce. Their offspring also enjoy a clear nomenclature. Li-ligers, li-tigons, ti-ligers, and ti-tigons are all readily bred from female ligers and tigons, and the female offspring of these crosses are also fertile. P. concolor × L. pardalis (puma × ocelot) bridges the gap between larger and smaller cats.27 Hybridization data connects the largest cats, P. tigris and the massive Liger (400+kg), to the smallest cat, F. nigripes, via seven documented hybrid steps: P. tigris (110–320 kg) × P. leo (120–250 kg) × P. pardus (30–85 kg) × P. concolor (35–100 kg) × L. pardalis (11–16 kg) × L. wiedii (3–9 kg) × F. catus (3–7 kg) × F. nigripes (1.5–2.5 kg). Intralineage hybrids of the lynx always involve the bobcat (L. rufus) and are referred to as Blynx or Lynx cats. Five cat hybrids are bred commercially as pets: F. chaus × F. silvestris (Euro-chaus), L. rufus × F. chaus (Jungle lynx), L. rufus × F. catus (desert lynx), F. catus × P. bengalensis (Bengal cat), and Bengal cat × F. chaus (Jungle bob).
Feline-specific viruses
Viruses can only reproduce with the help of host cells. Viruses first attach to specific surface molecules on the host cell; then they infect the cell. Some viruses tend not to be host-specific and are able to infect different species. A typical example of this is the influenza virus. It infects different families of birds, as well as pigs, other animals and, of course, man. The viruses attach to surface molecules common to warm-blooded animals in general. Other viruses, however, are far more host-specific. Feline Immunodeficiency Virus (FIV), the feline equivalent of HIV, has been shown to infect 30 of 37 felid species. At least four FIV strains are species-specific. However, it also infects Crocuta crocuta (the spotted hyena) a member of the felids’ taxonomically closest family. Nevertheless, the studies confirm that FIV transfer, even between cat species, is an infrequent event.28 The deadly viral disease, Feline Infectious Peritonitis (FIP), is specific to cats. Its causative agent, Feline Corona Virus (FCoV), is very similar to the human respiratory virus responsible for SARS and to coronaviruses from other animals. The mutation rate of the virus is high (three mutations per virus genome per generation). Since the disease was first clinically described, in 1963, no other natural host, besides members of the cat family, has been reported. There was a single report that FCoV could induce infectious peritonitis in ferrets,29 and dogs can be infected under laboratory conditions.30 Otherwise, FCoV is an infallible indicator of the felid basic type, from house cats to lions, with cheetahs being especially susceptible.31 Currently, host specificity of viruses is not considered diagnostic of a basic type. Nevertheless, these are additional pieces of evidence supporting the unique character of the feline basic type because, essentially, cats alone are susceptible, with rare exceptions.
Basic-type or holobaramin?
In 1998 an article about the family of cats was printed in the journal Creation Research Society Quarterly.32 The authors placed the family of cats within a so-called ‘holobaramin’. A holobaramin, as originally defined, is ‘a complete set of organisms’ that are genetically related to each other through common descent.33 Because common descent is harder to assess the further back in time one goes, eventually becoming empirically impossible to validate, Wood et al.34 proposed an alternative definition, “a group of known organisms that share continuity (i.e. each member is continuous with at least one other member) and are bounded by discontinuity”. To delimit baramin (a composite term derived from the Hebrew: bara = created, and min = kind), a modification of numerical taxonomy is used. In their article, Robinson & Cavanaugh33 examined as many characteristics from cats and closely related animals as possible. Characteristics included information from ecology, morphology, and genetic evidence, such as size, weight, proportion of bone lengths to each other, food, chromosome numbers, etc., to name a few. In total the authors compared 287 criteria from cats, Crocuta crocuta (the spotted hyena), and Macaca fascicularis (the long-tailed monkey). The proportion of characteristics not shared between two species was mathematically transformed into a dissimilarity value: the baraminic distance. For example, the distance between the two lynx species, Lynx lynx (Eurasian lynx) and L. canadensis (Canadian lynx) was 1.5%. Based on the characteristics examined, these animals are very similar to each other. The largest difference recorded in this analysis, 58.8%, was between Panthera leo (lion) and M. fascicularis (long-tailed monkey). Even so, some values are problematic; e.g. the distance between Caracal caracal (caracal) and P. leo (lion) was 36.4%, but the difference between P. onca (jaguar) and Cr. crocuta (spotted hyena) was only 35.9%. Treatment of such values calls for best-judgment decisions. Using a synopsis of all the values, the authors hypothesized that the family of cats represent a single holobaramin.
Over the years the two taxonomic concepts used to delineate a related group of species, the basic type and the baramin, have been converging. Even in their 1998 article, Robinson & Cavanaugh33 used hybridization data as an indicator of inclusion in a holobaramin. They wrote, “The potential for interspecific hybridization provides an important data set for elucidating monobaramins.”35 Further, Wood et al. wrote in their article about the HybriDatabase: “Within-group reproductive viability and outgroup reproductive isolation have been hypothesized to be important characteristics of the holobaramin or basic type.”36 The more the hybridization criterion is given primary significance, the closer the application of the holobaramin concept approaches the basic type definition. In turn, basic type research requires supplementary methods, such as statistical tools, to place species that cannot be crossed, as is clearly the case with fossils. In summary, it can be stated that the family of cats represents both a basic type and a holobaramin according to external characteristics, hybridization data, and genetic evidence.
Conclusion
Ability to hybridize is the most important criteria for including species within a common basic type.37 This criterion cannot be used directly on fossil forms. However, because it indicates the extent of the morphogenetic potential of a basic type, hybridization is an indirect indicator of fossil inclusion. Therefore, reports of extant cat hybrids, fossil skeletal evidence, and various other features, including molecular sequencing data, pelage patterns, and unique virus sensitivities, all seem to suggest that the family Felidae represents a single clearly delineated basic type. It is reasonable to assume that all felids arose from a single founder species and that they have passed through one or more adaptive radiations, exploiting their inherent morphogenetic potential to produce all of the known extant and extinct species of cat.1
References and notes
- Crompton, N.E.A., The Feline Basic Type, Tagungsband der Fachtagung für Biologie, SG Wort und Wissen 15:9–15, 1998. Return to text.
- Kitchener, A.C., Beaumont, M.A. and Richardson, D., Geographical variation in the clouded leopard, Neofelis nebulosa, reveals two species, Curr. Biol. 16:2377–2383, 2006. Return to text.
- Alderton, D., Wild cats of the world, Blanford books, London, 1993. Return to text.
- Macdonald, D., The velvet claw: a natural history of the carnivores, BBC Books, BBC Enterprises Ltd, London, 1992. Return to text.
- Martin, L., Felidae; in: Janis, C.N., Scott, K.M. and Jacobs, L.L. (Eds.), Evolution of Tertiary Mammals of North America, Cambridge University Press, Cambridge, pp. 236–242, 1998. Return to text.
- Turner, A. and Anton, M., The big cats and their fossil relatives, Columbia University Press, New York, 1997. Return to text.
- Martin, L., Nimravidae; in: Janis, C.N., Scott, K.M. and Jacobs, L.L. (Eds.), Evolution of Tertiary Mammals of North America, Cambridge University Press, Cambridge, pp. 228–235, 1998. Return to text.
- Kitchener, A., The Natural History of the Wild Cats, Cornell University Press, New York, 1991. Return to text.
- Flynn, J.M. and Galiano, H., Phylogeny of early Tertiary Carnivora, with a description of a new species of Protictis from the middle of the Eocene of Northwestern Wyoming, American Museum Noviates 2725:1–64, 1982. Return to text.
- Neff, N., The big cats: the paintings of Guy Cohelaech, New York, 1983. Return to text.
- Owen, R., On the anatomy of the cheetah Felis jubata, Schreb. Trans. Zool. Soc. London 1:129–136, 1835. Return to text.
- Peters, G. and Hast, M.H., Hyoid structure, laryngeal anatomy, and vocalization in the Felids (Mammalia: Carnivora: Felidae), Zeitschrift für Säugetierkunde 59:87–104, 1994. Return to text.
- Weissengruber, G.E., Forstenpointer, G., Peters, G., Kübber-Heiss, A. and Fitch, W.T., Hyoid apparatus and pharynx in the lion (Panthera leo), jaguar (Panthera onca), tiger (Panthera tigris), cheetah (Acinonyx jubatus) and the domestic cat (Felis silvestris f. catus), J. Anat. 201:195–209, 2002. Return to text.
- Wing, S.L., Tertiary vegetation of North America as a context for mammalian evolution; in: Janis, C.N., Scott, K.M. and Jacobs, L.L. (Eds.), Evolution of Tertiary Mammals of North America, Cambridge University Press, Cambridge, pp. 37–65, 1998. Return to text.
- Janis, C.N., Scott, K.M. and Jacobs, L.L., Evolution of Tertiary Mammals of North America, Cambridge University Press, Cambridge, 1998. Return to text.
- Weigel, I., Das Fellmuster der wildlebenden Katzenarten und der Hauskatzen in vergleichender und stammesgeschichtlicher Hinsicht, Säugertierk. Mitt. 9:1–120, 1961. Return to text.
- Werdelin, L. and Olsson, L., How the leopard got its spots: a phylogenetic view of the evolution of felid coat patterns, Biol. J. Linnean Soc. 62:383–400, 1997. Return to text.
- Liu, R.T., Liaw, S.S. and Maini, P.K., Two-stage Turing model for generating pigment patterns on the leopard and the jaguar, Phys. Rev. 74 E:011914, 2006. Return to text.
- Eizirik, E., Yuhli, N., Johnson, W.E., Menotti-Raymond, M., Hannah, S.S. and O’Brien, S.J., Molecular genetics and evolution of melanism in the cat family, Curr. Biol. 13:448–453, 2003. Return to text.
- Kim, J.-H., Antunes, A., Luo, S.-J., Menninger, J., Nash, W.G., O’Brien, S.J., and Johnson, W.E., Evolutionary analysis of a large mtDNA translocation (numt) into the nuclear genome of the Panthera genus species, Gene 366:292–302, 2006. Return to text.
- Christiansen, P., Pylogeny of the great cats (Felidae: Pantherinae), and the influence of fossil taxa and missing characters, Cladistics 24:977–992, 2008. Return to text.
- Johnson, W.E., Eizirik, E., Pecon-Slattery, J., Murphy, W.J., Antunes, A., Teeling, E. and O’Brien, S.J., The late Miocene radiation of modern Felidae: a genetic assessment, Science 311(5757):73–77, 2006 | doi:10.1126/science.1122277. Return to text.
- Lumpkin, S., Small Cats Facts on File, New York, 1993. Return to text.
- Trut, L.N., Plyusnina, I.Z. and Oskina, I.N., An experiment on fox domestication and debatable issues of evolution of the dog, Russian J. Genet. 40:644–655, 2004. Return to text.
- Driscoll, C.A., Menotti-Raymond, M., Roca, A., Hupe, K., Johnson, W.E., Geffen, E., Harley, E.H., Delibes, M., Pontier, D., Kitchener, A.C., Yamaguchi, N., O’Brien, S.J. and Macdonald, D., The Near Eastern origin of cat domestication, Science 317(5837):519–523, 2007 | doi: 10.1126/science.1139518. Return to text.
- Gray, A.P., Mammalian hybrids, Commonwealth Agricultural Bureau, Slough, UK, 1972. Return to text.
- Dubost, G. and Royere, J.-Y., Hybridization between ocelot (Felis pardalis) and puma (Felis concolor), Zoo Biol. 12:277–283, 1992. Return to text.
- Troyer J.L., Pecon-Slattery, J., Roelke, M.E., Johnson, W., VandeWoude, S., Vazquez-Salat, N., Brown, M., Frank, L., Woodroffe, R., Winterbach, C., Winterbach, H., Hemson, G., Bush, M., Alexander, K.A., Revilla, E. and O’Brien, S.J., Seroprevalence and genomic divergence of circulating strains of feline immunodefi ciency virus among Felidae and Hyaenidae species, J. Virol. 79:8282–8294, 2005. Return to text.
- Martinez, J., Ramis, A.J., Reinacher, M. and Perpinan, D., Detection of feline infectious peritonitis virus-like antigen in ferrets, Vet. Record 158:523, 2006. Return to text.
- Pedersen, N.C., A review of feline infectious peritonitis virus infection: 1963–2008, J. Feline Med. Surg. 11:225–258, 2009. Return to text.
- Kennedy, M., Citino, S., McNabb, A.H., Moffatt, A.S., Gertz, K. and Kania, S., Detection of feline coronavirus in captive Felidae in the USA, J. Vet. Diagn. Invest. 14:520–522, 2002. Return to text.
- Robinson, D.A. and Cavanaugh, D.P., Evidence for a holobaraminic origin of the cats, Creation Res. Soc. Quart. 35:2–14, 1998. Return to text.
- ReMine, R.J., Discontinuity systematics: A new methodology of biosystematics relevant to the creation model; in: Walsh, R.E. and Brooks, C.L. (Eds.), Proceedings of the Second International Conference on Creationism, Creation Science Fellowship, Pittsburgh, PA, pp. 207–213, 1990. Return to text.
- Wood, T.C., Wise, K.P., Sanders, R. and Doran, N., A refined baramin concept, Occasional Papers of the BSG 3:1–14, 2003. Return to text.
- Defined in Wood, ref 35, et al. as “a group of known organisms that share continuity, without regard to discontinuity with other organisms”. Return to text.
- Wood, T.C., Wise, K.P., Mace, S., Ingolfsland, K., Brown, M., Burleson, J., Celius, T., Clark, M., Clemons, J., Dahlke, D., Drake, S.K., Evans, D., Gonce, E., Haase, D., Hall, J., Heathershaw, P., Hughes, J., Hughes, K., Joines, C., Magill, T., Martin, S., Mitchell, J., Norquist, B., Smith, E., Snavely, C., Storie, J., Wasser, J., Woodlee, A., Kreps, J.L. and Robinson,D.A., HybriDatabase: A computer repository of organismal hybridization data; in: Helder, M.J. (Ed.), Discontinuity: Understanding Biology in the Light of Creation, Baraminology Study Group, p. 30, 2001. Return to text.
- Scherer, S., Basic types of life; in: Scherer, S. (Ed.), Typen des Lebens, Pascal Verlag, Berlin, 1993. Return to text.




Readers’ comments
Comments are automatically closed 14 days after publication.