Taking a crack at the Neandertal mitochondrial genome

The recent publication of a full-length Neandertal mitochondrial genome has been generating a lot of interest.1 The paper concluded that Neandertals are outside the range of modern human variation, and are therefore something different. This is a challenge to the standard creationist position that both Neandertals and modern humans are descended from the eight people who survived the Flood. Can we still include Neandertals in the human family tree? Are they descended from Noah? Does this single Neandertal sequence invalidate the Genesis accounts of Creation, the Flood, or the Tower of Babel?
First, an analysis of the authors’ methodology is beyond the scope of this article. For the sake of argument, let us assume that the authors were careful and that their techniques are sufficient for generating what they claim to have found. (There exists a large body of literature dealing with the pitfalls and assumptions inherent in working with ancient DNA. We should be skeptical of evolutionary studies, but sometimes we have to take things at face value for a time. The authors are aware of this knowledge and did their best to avoid potential problems. Only time will tell if their techniques lead to verifiable results.)
What did they find? They found that Neandertal mitochondrial2 DNA is different from that of modern humans. My calculations indicate this Neandertal specimen differs from the standard human mitochondrion (the Revised Cambridge Reference Sequence,3 or rCRS) by 210 nucleotides (1.3% of the 16,569 nucleotide mitochondrial genome). While 210 nucleotide differences sounds like a lot, it is not really fair to compare an ancient individual’s DNA with that of a modern person (the rCRS comes from a modern European). If one could back up the mutational history of the rCRS to the place where Neandertals and Europeans separated, one would find at least 12 fewer differences.4
That still leaves about 200 mutations separating the Neandertal specimen from modern humans. Is that a lot? To put that in perspective, any two modern humans selected at random will differ by an average of about 40 nucleotides. Although the most divergent mtDNAs from living humans differ at over 100 nucleotides, 200 mutations is still a lot, but it is not outside of reasonable probabilities.
I ran some quick calculations and discovered that only 60 of those 210 mutations are unique to Neandertal. The others are found in at least one other human within the available mitochondrial sequence databases. These mutations are pretty standard (no large indels, transversions are uncommon, etc.). The remaining mutations are all standard as well. There is nothing unusual about them, other than their number. But would we expect such an old sample to have so many mutations? Did not this individual live many generations ago and should we not expect it to have fewer mutations than people living today? Can we even tell which line has more mutations?
Mutation Rates are Unknown

To answer these questions, we must first understand that we know nothing about historic mutation rates and next to nothing about modern rates. Most mutation rate estimates we see in the scientific literature are biased downwards because of the assumption of deep-time evolution. This is a problem for two reasons. One, they may be calculating divergence times for two species that were created separately (e.g., chimps and humans) and are thus not technically comparable. Two, they divide the differences by a very large number. If humans and chimps are X% different, to calculate mutation rates, they divide X by 6 million (the number of years since we supposedly diverged from chimps). This always generates a low estimate of mutation rates. The timing of the split between modern humans and Neandertals is based on this very low rate calculated from the assumed human-chimp divergence time. Mutation rates based on genealogy are much higher than those based on phylogeny5 and are probably much more realistic. Recent studies have shown that measurable mutation rates are much higher than either the phylogenetic or even the genealogical methods predict.6
The Neutral Model of Evolution is Problematic
The Neandertal study also assumes the Standard Neutral Model of evolution7 and the presence of a molecular clock. Without going into too much detail, let us just say that the neutral model makes the assumption that mutation accumulation in mitochondria occurs without natural selection and that the genetic (and cultural) factors that control mutation rates do not vary across the human population or over time.
Thus, since we do not know actual mutation rates, since we do not know historic mutation rates, and since the assumptions behind the Standard Neutral Model are all questionable, the degree of relatedness of this single Neandertal specimen to modern humans is completely up in the air.
Neandertal Diversity is Unknown
Comparing a single Neandertal to all modern humans does not tell us much. When we know more about Neandertal genetic diversity, we will be able to make a better conclusion about how similar they are to modern humans. I suspect that Neandertal diversity is greater in general, but not because of some evolutionary reason. It is entirely possible that Neandertals accumulated mutations very rapidly in the years after the Flood. By way of comparison, there is a huge amount of diversity among living chimpanzees,8 perhaps as much as three to four times as much diversity as within the entire human population.9 This single Neandertal sequence is closer to modern humans than many living chimps are to one another!
Coalescence and Genetic Drift

Mathematically, lineages “coalesce” over time in small populations. That is, if you were to build a family tree for a small population, you would probably find a common maternal and paternal ancestor for the group just by chance. This is the reason why the survivors of the Mutiny on the Bounty, living today on Pitcairn Island, all have the same last name (see No Bones About Eve)! Because women generally take on the last name of their husband (especially in the past), last names are an excellent substitute for tracking the Y chromosome, which is passed on from father to son. Females do not count, for they do not inherit the Y chromosome and they get to change their name. But females have their own version of a last name, the mitochondrial DNA, which passed from mother to child. Men have the same mitochondrial DNA as their mother, but they do not pass it on to their children. Therefore, it is possible to build a family tree for both Y chromosomes and mitochondrial DNA. The Out of Africa Theory10 originally postulated that all people alive today descend from a single female (“Mitochondrial Eve”) living in Africa about 200,000 years ago (the estimated date varies from author to author). This does not mean that she was the only female alive at that time, but that the lineages of every living person coalesce in this single person. The theory has since been expanded to include “Y Chromosome Adam”. I find it humorous that evolutionary theory claims that we all came from a single male and female ancestor (even though they do not claim that they were necessarily living at the same time).
What does coalescence have to do with Neandertals? If we were to go back in imaginary evolution time to the point where modern humans and Neandertals supposedly split off from one another, you would find a population of proto-humans. Because coalescence is more or less random, it would be unlikely for the direct ancestor of both modern humans and Neandertals to be the same person within that population. The modern human and Neandertal populations each derived from a small subset of the people alive at the time, so the respective family trees started with different genetic lineages. Therefore, according to theory, one would not expect to find many mutations in common among modern humans and ancient Neandertals. We came from different genetic stock to begin with. The “random sampling” nature of coalescence predicts disparity. But the YEC position does not depend on this line of reasoning. In our model, the early post-Flood human population of about 1,000-10,000 people was split apart at Babel. Different groups went different directions on the globe and each saw different environmental pressures, experienced different demographic pressures, and were subject to different mutational events. Neandertals are one of those groups. They are different because they had a different history. We do not yet know much about that history, but we are learning more with time.
Inbreeding
Genetic drift occurs more rapidly in small populations. According to standard population genetics theory, the likelihood of any new mutation displacing all other alleles within the population is directly proportional to the size of the population. That is, in a population of 1000 individuals, a new mutation has a chance of 1/1000 of becoming “fixed”. If the Neandertal population was small or was restricted to a small size for a number of generations (as Green et al.1 hypothesize), genetic drift could have occurred rapidly.
Small populations are also at a greater risk of extinction due to the high rate of mutation accumulation,11 which eventually leads to extinction due to “error catastrophe”. This Neandertal sequence has many deleterious mutations, several of those in one particular gene.1 Why is this so? Should not natural selection have weeded out these negative mutations? Instead, they accumulated. The authors of the paper said that “purifying selection” had broken down. This is evidence for a high mutation rate acting on a small population. Exactly what happened to the Neandertals, anyway?
Rapid mutation in harsh environments
Bruce Ames, a member of the prestigious US National Academy of Science, has suggested that genetic damage can be directly linked to poor nutrition.12 According to the theory, when under starvation conditions, the body has to decide which systems to keep working and which to shut down. This genetic “triage” mechanism would keep an organism alive, but at the expense of less-than-critical cellular operations, like DNA repair.
It has been suggested by several creationists that the Neandertal population lived in Europe under less-than-ideal conditions and was subjected to nutrient limitations, specifically vitamin D deficiency due to the perpetually cloudy weather during the post-Flood Ice Age (see The Caring Neandertal and Bones Overthrown). Couple a harsh environment and poor nutrition to a small inbreeding population and you have an instant recipe for the rapid accumulation of mutations.
Predetermined mutation pathways?
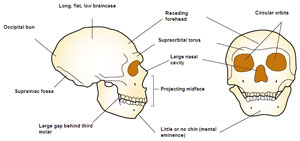
One of the assumptions behind neutral theory is that all mutations are independent and random. It is probably a mistake to believe that mutations occur at random and that they do not interact, however, for any mutation can only occur in the context of the surrounding genetic information. One mutation may be excluded by another (because the combination might be deadly) or may lead to a series of other mutations (because some mutation sets may be excluded by specific individual mutations). Evidence for this kind of mutation interaction is limited, but it does exist.5 If some mutations lead to others, we should not expect two separate lineages to follow the same mutational pathway. This is especially true if a mutation affects the fidelity of the DNA copying mechanism. Remember, neutral theory does not allow for mutations in DNA polymerase or in anything that affects DNA copying or repair. Too bad for Standard Neutral Model of evolution theory that such things are easily demonstrated.13,14
Conclusions
While evolutionists (including theistic evolutionists) and ‘progressive creationists’ will probably be trumpeting this new paper as evidence that Neandertals and modern humans are two distinct species, I believe their conclusions are premature. As I have briefly outlined above, there is a lot we do not know about the science of modern genetics. And there are factors like coalescence and genetic drift in small isolated populations that can potentially explain the findings. In any case, modern chimps can differ more from each other in their mtDNA than modern humans differ from this Neandertal specimen, so beware of anyone who claims Neandertals are a separate species based on genetic differences.
When we approach evidence like this, we need to be skeptical, we need to understand the theory that led to the conclusions, and we need to question the assumptions behind the theory. If we do these three things, we need not be afraid that Neandertal Man will in some way fall outside the biblical creation model.
References
- Green, R.E., Malaspinas, A.-S., Krause, J., Briggs, A.W., Johnson, P.L.F, Uhler, C., Meyer, M., Good, J.M., Maricic, T., Stenzel, U., Prüfer, K., Siebauer, M., Burbano, H.A., Ronan, M., Rothberg, J.M., Egholm, M., Rudan, P., Brajković, B., Kućan, Ž., Gušić, I., Wikström, M., Laakkonen, L., Kelso, J., Slatkin, M., and Pääbo, S. A Complete Neandertal Mitochondrial Genome Sequence Determined by High-Throughput Sequencing. Cell 134:416–426, 2008. Return to text.
- Different from the nuclear DNA, which constitutes most of the DNA within a cell. A mitochondrion is a small organelle within the cell which is like an ‘energy factory’. It has its own small store of DNA. Return to text.
- Andrews, R.M., Kubacka, I., Chinnery, P.F., Lightowlers, R.N., Turnbull, D.M., and Howell, N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nature Genetetics 23:147, 1999. Return to text.
- Carter, R., Mitochondrial diversity within modern human populations. Nucleic Acids Research 35(9):3039–3045, 2007. Return to text.
- Howell, N., Smejkal, C.B., Mackey, D.A., Chinnery, P.F., Turnbull, D.M., and Herrnstadt, C. 2003. The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates. American Journal of Human Genetics 72:659–670. Return to text.
- Elliott, H.R., Samuels, D.C., Eden, J.A., Relton, C.L., and Chinnery, P.F.. 2008. Pathogenic Mitochondrial DNA Mutations Are Common in the General Population. American Journal of Human Genetics 83:254–260. Return to text.
- Kimura, M., Evolutionary rate at the molecular level. Nature 217:624–626, 1968. See also Theory Of Molecular Evolution, wikipedia.org. Return to text.
- Becquet, C., Patterson, N., Stone, A.C., Przeworski, M., and Reich, D., Genetic Structure of Chimpanzee Populations. PLoS Genetics 3(4):617–626, 2007. See also Chimpanzees Are Actually Three Distinct Groups, Gene Study Shows. sciencedaily.com. Return to text.
- Kaessmann, H., Wiebe, V., and Pääbo, S. 1999. Extensive Nuclear DNA Sequence Diversity Among Chimpanzees. Science 286:1159–1162,, 1999. See also Chimpanzee Subspecies Are Genetically Mixed And More Diverse Than Humans, sciencedaily.com. Return to text.
- Cann R.L., Stoneking, M., and Wilson, A.C., Mitochondrial DNA and human evolution. Nature 325:31–36, 1987. Return to text.
- Lynch, M., Conery, J., and Burger, R., Mutation accumulation and the extinction of small populations. American Naturalist 146:489–518, 1995. Return to text.
- Ames, B., Low micronutrient intake may accelerate the degenerative diseases of aging through allocation of scarce micronutrients by triage. PNAS (USA) 103(47):17589–17594, 2006. Return to text.
- Copeland, W.C., Ponamarev, M.V., Nguyen, D., Kunkel, T.A., and Longley, M.J., Mutations in DNA polymerase gamma cause error prone DNA synthesis in human mitochondrial disorders. Acta Biochemica Polonica 50(1):155–167, 2003. Return to text.
- Lee, H.R., and Johnson, K.A., Fidelity of the Human Mitochondrial DNA Polymerase. The Journal of Biological Chemistry 281(47):36236–36240, 2006. Return to text.



Readers’ comments
Comments are automatically closed 14 days after publication.