Fast folding cells perform a truly amazing feat … and have from the beginning
One of the biggest mysteries in biology is how the body’s cells fold up long protein molecules.
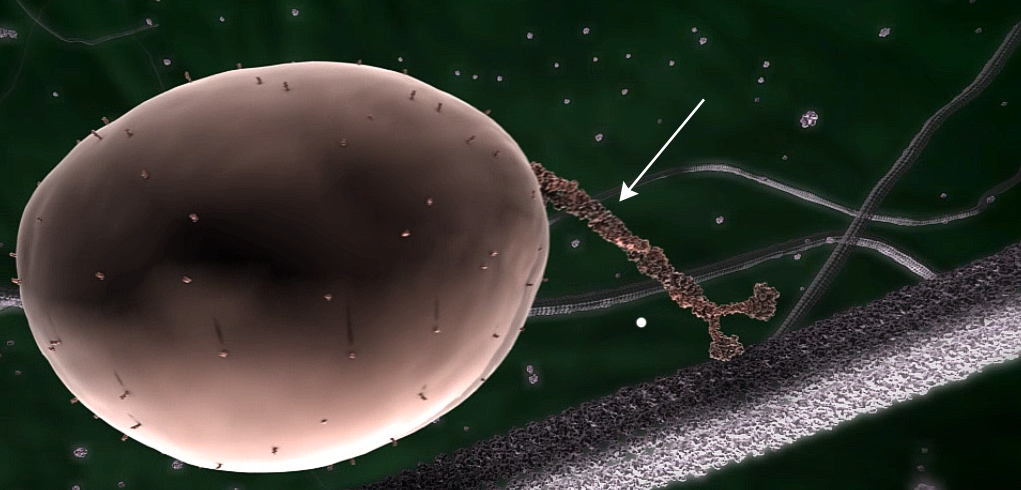
All living things have proteins in their cells, each made up of a long chain of amino acids in a particular sequence. This sequence is specified by the DNA ‘instructions’ for making it. This chain is then folded to enable it to perform its particular function, using previously folded proteins called chaperonins. For example, the complex biomolecular machines that transport substances within cells, called kinesins, are made up of such chains, folded into a three-dimensional structure that gives each one its unique structure and function (fig. 1).
Individual amino acids must bond with certain other amino acids at various parts of the chain in particular ways during the stages of folding. The task is astronomically difficult and incredibly complex—yet cells can do it in less than the blink of an eye. Cells also need to quickly unfold the protein chains, too.
But how does life get the atomic-precision-level 3D shapes from a 1D ‘letter’ sequence (a one-dimensional string of amino acids)? Scientists have been working for decades trying to predict the shape from the sequence. But the problem was too difficult for even the world’s most powerful classical supercomputers to simulate. However, in late 2020, Google’s DeepMind program AlphaFold used artificial intelligence trained on an extensive protein structure database to solve protein structures.1
But what about solving from scratch? Classical computing uses an approach that tries every possible solution. But as IBM research scientist and paper co-author2 Panagiotis Barkoutsos explains, folding is an exponential problem.3 As each additional bond occurs the chain becomes more and more difficult to fold. Soon it becomes almost impossibly complex. In 1969, molecular biologist Cyrus Levinthal estimated that if cells took this ‘try every possibility’ approach they would take longer than the billions-of-years claimed evolutionary age of the universe to complete one fold. This is known as Levinthal’s Paradox.
It has long been hoped that such problems could be solved one day with the advent of quantum computing, still in its infancy. Rather than using the ‘on/off’ (or ‘yes/no’) logic switches of classical computing, quantum computers function by what is called superposition—making use of probability—where something is both on and off at the same time. That means that they should be able to perform in a tangible timeframe some types of calculations that would otherwise take millions of years.
In 2021, the IBM Quantum, a computer at the cutting edge of this new technology, managed to simulate the folding of a protein. It used 22 quantum bits (or qubits). These are analogous to a ‘bit’ in ordinary computing; adding a qubit doubles the processing power, so 22 of them are 4 million times faster than a single qubit.
For proteins (and it would work for other large molecules), the simulation superimposed all physically meaningful folding combinations. Then the program statistically sampled combinations to see which would result in the lowest energy state. This would reflect the real world where molecules move to lower energy states, the most stable.
This feat involved a very small protein with only 10 amino acids, called angiotensin, which constricts blood vessels and elevates blood pressure. The simulation managed to fold this into a very simple triangular pyramid form. (Another simulation needed only 9 qubits of a 20-qubit quantum computer to solve the structure of a nerve protein 7 amino acids long.) However, most protein chains are hundreds or thousands of amino acids long. And in real life they must fold into much more complex, specific three-dimensional shapes to perform their particular biological function.
The design of kinesins, involving in effect programmed coded instructions enabling them to perform their tasks, is amazingly complex. If protein folding doesn’t occur, life can’t exist. But the DNA and other machinery guiding the folding needs already-folded proteins in order to exist and operate. This is a classic ‘chicken-and-egg’ conundrum for evolution. Living things must have been able to perform this incredible feat from the moment they were created.
References and notes
- Callaway, E., ‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures: Google’s deep-learning program for determining the 3D shapes of proteins stands to transform biology, say scientists, nature.com, 30 Nov 2020. Return to text.
- Robert, A. and 3 others, Resource-efficient quantum algorithm for protein folding, NPJ Quantum Information 7:38, nature.com, 17 Feb 2021. Return to text.
- Letzter, R., A novel quantum algorithm for protein-folding: paving the way toward resolving one of the biggest mysteries in biology with quantum computers, medium.com, 20 Aug 2021. Return to text.








Readers’ comments
Comments are automatically closed 14 days after publication.