Journal of Creation 28(3):5–8, December 2014
Browse our latest digital issue Subscribe
Denisovans menace evolution—a new chapter in the human origins debate
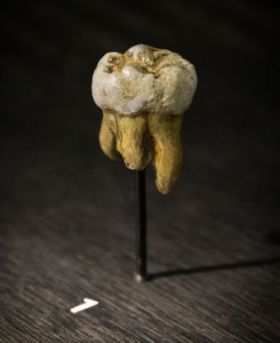
A new chapter in the human origins debate opened in the year 2010 with the discovery of a new kind of archaic human called Denisova. Now not just the fossils are available to researchers but also DNA. Paleogenetics can now allegedly settle long-lasting questions due to the incompleteness of the fossil record, although DNA sequence veracity is a matter of concern among creationists.1
Denisovans were discovered in the Upper Paleolithic layer 11.1 of Denisova Cave in southern Siberia, their remains consisting of, surprisingly, a distal manual phalanx and a molar tooth found at the same archeological site from two individuals supposedly from the same population.2
The Denisovan genome has been analyzed over the past few years, with sweeping claims of their cognitive capabilities, external appearance, and even detailed population dynamics. Based on such a small number of fossil remains, it is premature to draw too many robust scientific conclusions from the analysis of Denisova. Creation theory would predict that an archaic human would fit very well into the created human kind, as we shall see in the following.3
Genetic contributions of Denisova to the human genome
According to various estimates, Denisovans are also thought to have contributed to some 2–7% of the genetic material of Melanesians, Australian Aborigines, and other Southeast Asian islanders.4,5,6 Evolutionists have attempted to force fit Denisovans into their evolutionary models, which state that Denisovans, along with Neandertals supposedly broke off 400,000 years ago from the common ancestors with modern humans. Denisovans seemingly also interbred with Neandertals, as a recent study showed this year, which determined an almost complete mitochondrial genome sequence taken from a femur bone found at a site called Sima de los Huesos (‘pit of bones’ in Spanish).7 The femur, as well as the other bones at the site, is attributed to Neandertals, yet the mitochondrial genome sequence itself more resembles Denisova. Denisovans were first discovered in southern Siberia, thus a menacing question for evolutionists is how Denisovans could possibly widely overlap Neandertals geographically (from Spain to Oceania), when they were supposed to be diverging as a new species. Denisovan genetic variety has also been shown to be quite large, being seven times that of Neandertals, based on their mitochondrial DNA.5,6
According to the biological definition of a species, two individuals belong to the same species if they can interbreed and thus share common genetic material. Denisovans contributed genes coding for the human leukocyte antigen system (HLA-A and HLA-C) to Melanesians, Asians, and Europeans, which help the immune system fight pathogens, as well as the KIR3DS1*013 allele (killer-cell immunoglobulin-like receptor).8 Reich et al.9 theorize on how Denisovans contributed genetic material to Southeast Asians, Papuans, Australians, and Pacific Islanders as ancient humans migrated throughout Southeast Asia and Oceania. Other researchers found that the transcription factor EPAS1 in Tibetans is likely to have originated from Denisovans, and Han Chinese to a smaller degree. This transcription factor is induced under hypoxic conditions, leading to higher quantities of hemoglobin in the blood in oxygen-scarce conditions; such oxygenation is needed in the high mountains of Tibet. SNP variants in a 32.7-kbp region show that a distinct five-SNP motif, AGGAA, is almost unique to Tibetans (it appears in some Han Chinese), among 26 populations from the Human Genome Diversity Panel, and matches exactly the Denisovan haplotype in this region.10
Evolutionists point out differences between the human and Denisovan genome in an attempt to explain how these two lineages evolved separately from one another. Overall, there are 23 single-nucleotide changes (SNCs) that have supposedly accrued on the lineage purported to be specific to Denisova which affect amino acid changes in certain proteins. Eight of these are related to brain function and nervous system development. Two of these genes have been linked to autism and language disorders (ADSL and CNTNAP2). Overall, 34 genetic variants in the Denisovan genome have been clearly associated with genes involved in human disease, such as EVC2, which causes Ellis-van Creveld syndrome. Taurodontism is also associated with this disease, and is suspected in the Denisovan molar found with fused roots and a large dental pulp cavity.

It is significant that no new genes had been found in Denisova, only the previously mentioned 34 new gene variants (some listed in table 1), the protein sequence of which differs from the human protein sequence generally in only 1–2 amino acids, which cause diseases. This is what we would expect if Denisovans and humans both belonged to the same created kind. The question, however, begs itself as to whether it is realistic to expect that all of these genetic variants simultaneously acquired advantageous mutations on the lineage leading to humans, as evolution would have it. These genes all cause illnesses due to base substitutions, thus it is more plausible that these genes underwent detrimental mutations, causing illness in Denisovans, who are now extinct, anyway.
Interestingly enough, both the FOXP2 gene, which is associated with the emergence of modern language, and the KLK8 gene, which is preferentially expressed in the central nervous system and is involved in learning and memory,11 have the same sequence in Denisova, Neandertal, and human, showing that they all had the same type of higher cognitive functions. Also noteworthy are so-called de novo genes, which supposedly arose through random base mutations from transcriptionally active, noncoding loci. Taylor12 examined the distribution of 60 such supposed genes in Neandertals, Denisovans, and modern humans. One such gene, CLLU1, underwent a single base mutation (ΔA), and thus supposedly became active in the Neandertal lineage. This would have all been well for evolution, except for the fact that this ΔA ‘enabler’ mutation is also missing from several southern African individuals; thus it is not specific to a supposed separate Neandertal evolutionary lineage.
Another interesting topic is the study of olfaction in modern humans compared to Denisovans and Neandertals. An analysis of the cranium of modern humans and Neandertals shows a large difference in size of olfactory bulb. However, the size of the olfactory bulb also differs between smokers and non-smokers in modern human populations. Twenty olfactory receptor (OR) genes were compared between modern humans, Neandertals, and Denisovans.13 Several combinations of these genes were lost in each lineage, according to adaptation to certain ecological niches (10 in Neandertals and 8 in Denisovans). This is in accordance with the creation model, which accurately predicts degradation of the genome over time. Neandertals and Denisova could have lost these OR genes in the cold climate during the post-Flood Ice Age, as lower temperatures do not favour odour volatility.
Common retrovirus insertions

According to Agoni et al.,14 insertions of 14 Endogenous Retrovirus (ERV/HERV-K) loci were supposedly found to be common only between Denisovans and Neandertals and not in humans, suggesting that these two species have an evolutionary lineage separate from modern humans. Endogenous retroviruses are common and make up about 5% of the human genome. However, many of the loci for these ERVs have been found by Marchi et al.15 in a number of new modern human genomes (table 2). For 8 of the 14 loci with exact genomic locations (the others were found inside repeat regions such as Alu sequences), seven were found in 67 cancer patients and 43 in other human genomes. Since the other loci occur within repeat regions, evidence for their existence in the modern human genome debunks the original claim that the variability for a small subset of these loci was limited to only archaic humans.
A newly discovered type of ERV/HERV-K, termed K111, was analyzed in the centromeric regions of chromosomes from 189 human DNA samples. In humans, K111 was found in hundreds of copies in the centromeres of 15 chromosomes. Reads for seven K111 insertions were found in Neandertal and four in Denisova.16 It is likely that K111 was found in such low numbers due to the difficulties in sequencing archaic genomes as well as their low coverage—combined with the difficulty of computationally assembling DNA sequences obtained from repetitive regions. Thus, as was the case with the previously mentioned HERV-K sequences, K111 is not specific to a supposed evolutionary linage either.
Chromosomal similarities
From a karyotypic viewpoint, great apes have 24 sets of chromosomes, while humans have 23. According to evolutionary theory, two acrocentric chromosomes fused to form human chromosome 2 in a head-to-head orientation. Bergman and Tomkins17,18 studied the 798 bp region encoding the alleged fusion site on human chromosome 2 and used it in a BL ASTN query against the chimpanzee genome where it had no hits on the short arms of chimpanzee chromosomes 2A and 2B—the supposed sites of fusion origins. In addition, Tomkins19 later discovered, in 2013, that the alleged 798-base fusion is actually a functional second promoter region in the DDX11L2 gene’s first intron (transcribed on the minus strand)—playing an important role in alternative transcription of the gene. Of interest to the issue of Denisovans, as noted by Meyer et al. in 2012,2 is the hexamer sequence GGGGTT found in the alleged fusion site—a motif that functionally plays a role in transcription factor binding. In the genomic comparison of Meyer et al.,2 the motif was found 15 times in human and 12 times in the DNA fragments of the Denisovan genome,2 while they are lacking from bonobos and chimps. Oddly, this indicates that the fusion site was more degraded in Denisovans than in modern humans (an intact telomere motif = TTAGGG). In addition, the DDX11L2 gene (containing the alleged fusion site), along with many other genes in a region of 614,000 bases surrounding the fusion site, is also completely missing in chimpanzee.19,20 Collectively, this further proves that humans and Denisovans belong to the same species. Furthermore, it is interesting to note that the human and Denisovan genome are essentially identical to each other on a large scale. And both species share the majority of duplicated genomic segments longer than 9 kbp.2
Highly correlated methylation patterns
The methylation patterns of the Denisovan and Neandertal genomes have also been indirectly analyzed, revealing further information on the global similarity of these two genomes with the human genome. After death, cytosine residues often deaminate (the amine group is lost to oxidation). Methylated cytosine becomes thymine, whereas unmethylated cytosine becomes uracil. Thus, we can assume that regions with higher proportions of C > T transitions were once methylated regions in ancient human genomes. These regions were compared to methylated regions taken from osteoblast cells in modern humans. The correlation between the methylation levels of CpGs in modern human genomes and the mean C > T ratio for Denisova was 0.989, and 0.981 for Neandertal. Furthermore, the reconstructed promoter regions of 3,804 housekeeping genes falls within the variation of 20 studied modern humans. Overall, approximately 99% of the Denisovan and Neandertal genomes show no significant methylation differences compared to the human genome.21 Hernando-Herraez22 compared methylation levels in 99,919 CpGs and 12,593 genes between human, chimpanzee, bonobo, gorilla, and orangutan. Only 22% of the CpGs showed no significant differences; about 9% of the CpGs and 745 genes had significant differences, and 2,500 genes had some methylation differences between human and chimpanzee. Thus, modern humans and Denisovans are distinctly separate from chimpanzee and other monkey species based on methylation patterns of selected similar DNA segments.
Evolutionists point to difference in methylation patterns among the three genomes for support of their model. There are differences in three HOXD genes, which affect limb development, the SREBP-1 gene, which affects lipid homeostasis, and the aryl hydrocarbon receptor (AHR) gene, which is a toxin receptor. Differential methylation patterns might indeed cause changes in the function of these genes, but this is entirely beside the point. These changes are in methylation patterns only; they give no indication of how the DNA sequences that they regulate evolved in the first place. Thus, these kinds of evolutionary explanations and reasoning are shallow. Finally, the differences in methylation patterns in genes associated with bone development could partially explain the variability between modern and archaic humans in some skeletal features since the DNA sequences are identical.21
Conclusion
The sensational construction of the Neandertal and Denisovan genomes has opened a window on the genetics of new members of the human kind. As can be expected, Denisovans and Neandertals are variants of the human kind, and also seemingly interbred with each other as well as modern humans. It has been shown that there is an anatomic gradation between humans and Neandertals based on skull similarities. Hence, the two belong to the same species.23 Since also the Denisovan genome is very similar to the Neandertal genome, by transitivity, Denisova must also be human. Similar karyotypes, extremely small differences in the whole genome sequence, global similarities in duplicated regions, similar insertions of retroviruses and very similar methylation patterns all are in support of this. Furthermore, Denisovans have also contributed several variants of known genes to modern humans, pointing to gene flow between the two sub-species. Gene flow means interbreeding, which is only possible if humans and Denisovans both belong to the same created kind.
Since Denisovan remains were found in layers dated to the Pleistocene era, and these layers contained artifacts such as jewelry and tools, we can hypothesize that they are members of the human kind that arose after the Flood, and that they had human intelligence, which is also supported by the presence of a couple of genes indicative of language capability. Interestingly, human artifacts reflective of the social and spiritual aspects of human life were found in Denisova Cave, such as decorations made of bone, mammoth tusk, ostrich egg, mollusk shell, as well as two fragments of a bracelet made of dark green chloritolite, which also shows evidence of having been polished on a flat and stable abrasive surface. Tools characteristic of the Upper Paleolithic were also found in the cave, including needles with drilled-out eyes, awls-borers, and pendants made of animal teeth.5,24 These human artifacts suggest that Denisova had human intelligence.
There are differences between Denisovans and modern humans, however. This is best seen in the disease-causing amino acid changes that have occurred in a number of genes in Denisova. However, no new genes arose during the shared history of these two members of the human kind.
References and notes
- Criswell, D., Neandertal DNA and Modern Humans, CRSQ 45:246–254, 2009. Return to text.
- Meyer, M., Kircher, M., Gansauge, M.T. et al., A high-coverage genome sequence from an archaic Denisovan individual, Science 338(6104):222–226, 2012 | doi: 10.1126/science.1224344. Return to text.
- Wieland, C. and Carter, R., Not the Flintstones—it’s the Denisovans, creation.com/denisovan, 2011. Return to text.
- Reich, D., Green, R.E., Kircher, M. et al., Genetic history of an archaic hominin group from Denisova Cave in Siberia, Nature 468(7327):1053–1060, 2010 | doi:10.1038/nature09710. Return to text.
- Gibbons, A., Who were the Denisovans?,Science 333(6046):1084–1087, 2011 | doi: 10.1126/science.333.6046.1084. Return to text.
- Lalueza-Fox, C. and Gilbert, M.T., Paleogenomics of archaic hominins, Curr Biol. 21(24):R1002–1009, 2011 | doi:10.1016/j.cub.2011.11.021. Return to text.
- Meyer, M., Fu, Q., Aximu-Petri, A. et al., A mitochondrial genome sequence of a hominin from Sima de los Huesos, Nature 505(7483):403–406, 2014 | doi:10.1038/nature12788. Return to text.
- Abi-Rached, L., Jobin, M.J., Kulkarni, S. et al., The shaping of modern human immune systems by multiregional admixture with archaic humans, Science 334(6052):89–94, 2011 | doi:10.1126/science.1209202. Return to text.
- Reich, D., Patterson, N., Kircher, M. et al., Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania, Am. J. Hum. Genet. 89(4):516–528, 2011 | doi: 10.1016/j.ajhg.2011.09.005. Return to text.
- Huerta-Sánchez, E., Jin, X., Asan, Bianba, Z. et al., Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA, Nature 512(7513):194–197, August 2014 | doi:10.1038/nature13408. Return to text.
- Paixão-Côrtes, V.R., Viscardi, L.H., Salzano, F.M., Hünemeier, T. and Bortolini, M.C., Homo sapiens, Homo neanderthalensis and the Denisova specimen: New insights on their evolutionary histories using whole genome comparisons, Genet. Mol. Biol. 35(4 [suppl]):904–911, 2012 | PMID: 23413113. Return to text.
- Taylor, J.S., Did Neanderthals and Denisovans have Our De Novo Genes?, J. Mol. Evol. 78(6):321–323, 2014 | doi: 10.1007/s00239-014-9628-x. Return to text.
- Hughes, G.M., Teeling, E.C. and Higgins, D.G., Loss of olfactory receptor function in hominin evolution, PLoS One 9(1):e84714, 2014 | doi: 10.1371/journal.pone.0084714. Return to text.
- Agoni, L., Golden, A., Guha, C. and Lenz, J., Neandertal and Denisovan retroviruses, Curr. Biol. 22(11):R437–438, 2012 | doi:10.1016/j.cub.2012.04.049 Return to text.
- Marchi, E., Kanapin, A., Byott, M., Magiorkinis, G. and Belshaw, R., Neanderthal and Denisovan retroviruses in modern humans, Curr Biol. 23(22):R994–995, 2013| doi: 10.1016/j.cub.2013.10.028. Return to text.
- Contreras-Galindo, R., Kaplan, M.H., He, S. et al., HIV infection reveals widespread expansion of novel centromeric human endogenous retroviruses, Genome Res. 23(9):1505–1513, 2013 | doi: 10.1101/gr.144303.112. Return to text.
- Tomkins, J. and Bergman, J., The chromosome 2 fusion model of human evolution—part 2: reanalysis of the genomic data, J. Creation 25(2):111–117, 2011; creation.com/chromosome-2-fusion-2. Return to text.
- Tomkins, J., Genome-Wide DNA Alignment Similarity (Identity) for 40,000 Chimpanzee DNA Sequences Queried against the Human Genome is 86–89%, Answers Research J. 4:233–241, 2011. Return to text.
- Tomkins, J., Alleged human chromosome 2 “Fusion Site” encodes an active DNA binding domain inside a complex and highly expressed gene—negating fusion, Answers Research J. 6:367–375, 2013. Return to text.
- Fan, Y. et al., Gene content and function of the ancestral chromosome fusion site in human chromosome 2q13-2q14.1 and paralogous regions, Genome Research 12:1663–1672, 2002. | PMID: 12421752. Return to text.
- Gokhman, D., Lavi, E., Prüfer, K. et al., Reconstructing the DNA methylation maps of the Neandertal and the Denisovan, Science 344(6183):523–527, 2014 | doi: 10.1126/science.1250368. Return to text.
- Hernando-Herraez, I., Prado-Martinez, J., Garg, P. et al., Dynamics of DNA methylation in recent human and great ape evolution, PLoS Genet 9(9):e1003763, 2013 | doi: 10.1371/journal.pgen.1003763. Return to text.
- Wieland, C., Making sense of ‘apeman’ claims, chap. 7; in: One Human Family: the Bible, science, race, and culture, Creation Book Publishers, 2014. Return to text.
- Derevianko, A.P., Shunkov, M.P. and Volkov, P.V., A Paleolithic bracelet from Denisova cave, Archaeology, Ethnology, & Anthropology of Eurasia 34(2):13–25, 2008 | doi:10.1016/j.aeae.2008.07.002. Return to text.








Readers’ comments
Comments are automatically closed 14 days after publication.