Journal of Creation 25(2):32–39, August 2011
Browse our latest digital issue Subscribe
Cladistics, evolution and the fossils
Cladistics is the premier method used for determining evolutionary relationships in biology. The results of cladistics analyses, tree diagrams called cladograms, are often used as demonstrations of evolution. Though cladistics was developed by and for evolutionists, it still fails to demonstrate evolution, let alone biological reality. Evolution is still typically seen as the theoretical justification for using cladistics in paleontology, so the conclusion of evolution merely begs the question. Cladograms only demonstrate a nested hierarchy of biological characters; they tell us nothing about what produced the pattern. Evolutionary cladistics also depicts a simplistic view of biological change and fails to deal with pleiotropy within organisms. These problems were recognized by some evolutionists over 30 years ago, but their criticisms largely fell on deaf ears, most likely because their comments were used as ammunition by creationists. Many problems of phylogenetic inference that cladistics claims to solve still remain largely unsolved, such as distinguishing between homology and homoplasy. Perhaps the largest problem, however, is the illusion of evolution that cladograms and the language used to describe them give to the public. They both create the illusion of a resolved genealogy despite some cladists’ disavowal of any strict genealogical connotations.
What is cladistics?
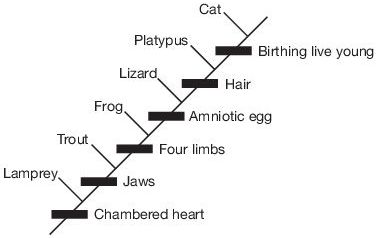
Cladistics has become the premier method that evolutionists use to map out evolutionary relationships in paleontology. Cladograms are ubiquitous in the paleontological literature, and are often used as evidence for evolution. Cladistics is a method that classifies organisms in a nested hierarchy of similarity based on a comparison of individual characteristics. It will identify a series of characteristics in each taxon for comparison (table 1), and then arrange the taxa in a cladogram (figure 1). Then different cladograms are compared in order to find which cladogram organizes the taxa in a hierarchy that has the least non-nested characters and/or the most nested characters. Evolutionists typically interpret the nested pattern as descent with modification. Character state changes are seen as phylogenetic changes.
History of cladistics
Cladistics was first proposed by Willi Hennig in 1950 as an alternative to then current systematic methods.1 However, Hennig did not coin the term ‘cladistics’, but preferred to call his method ‘phylogenetic systematics’, as he believed his method was a more empirically based way of constructing phylogenies. Rather, Ernst Mayr, a noted critic of Hennig, first coined the term ‘cladistics’ in 1965.2 Moreover, it wasn’t until 1966, when Hennig’s original work was revised and translated into English,3 that cladistics begun to have a substantial impact on English-speaking evolutionists.
Hennig argued that methods current in his day had two fundamental flaws: they were hopelessly subjective, and they failed to properly identify evolutionary relationships. Traditional Darwinian taxonomy was generally driven by the intuition of the individual biologist, which of course creates problems when disagreements arise because there are no evidential reasons to choose between the two. Phenetics sought to get around this by subjecting characters of organisms to pairwise comparisons, and thus evaluating the overall similarity between taxa. However, such a method would seem ill-equipped to deal with taxa that have similar forms, but are clearly not closely related, e.g. sharks, ichthyosaurs and dolphins. Character weighting then became inherent in the analysis, and it thus became as subjective as traditional Darwinian taxonomy.
Hennig countered that cladistics was able to identify homology empirically by identifying what he called synapomorphies, or ‘shared derived characters’. A comparison of individual traits across a range of taxa would reveal these synapomorphies, which he assumed arose through common ancestry.4
Evolutionary fights
Much of the focus then turned to the philosophical underpinnings of cladistics. By the 1980s most systematists agreed that cladistics was a useful methodology. However, there was considerable debate over what cladistics meant, and what it was supposed to be used for.
The dominant school of thought traced itself back to Hennig, and continued to argue that evolution is a necessary assumption for cladistics to work.5 The Hennigians were clear when they call cladistics ‘phylogenetic systematics’—they worked with the assumption that evolution is the foundation of cladistics. Therefore, they believed the purpose of cladistics was to elucidate the most probable evolutionary relationships that unfolded throughout history. Essentially, cladistics became an exercise in evolutionary theorizing.
Chambered | Jaws | Four | Amniotic | Hair | Birthing | |
|---|---|---|---|---|---|---|
Sea lamprey |
1 |
0 |
0 |
0 |
0 |
0 |
Rainbow trout |
1 |
1 |
0 |
0 |
0 |
0 |
Australian green tree frog |
1 |
1 |
1 |
0 |
0 |
0 |
Frill-necked lizard |
1 |
1 |
1 |
1 |
0 |
0 |
Platypus |
1 |
1 |
1 |
1 |
1 |
0 |
Cat |
1 |
1 |
1 |
1 |
1 |
1 |
Table 1. A simple cladistic analysis of character traits commonly held to be shared derived characters in vertebrates. Traits are polarized: 0–Absent and 1–Present.
However, some systematists broke with Hennig’s insistence that cladistics necessarily demonstrated transformation through character state changes.6 These so-called ‘transformed’ or ‘pattern’ cladists called this process assumption of Hennigian cladistics into question by saying that the methodology does not require the assumption of evolution to work. For transformed cladists, the purpose of cladistics was classification based on a descriptive definition of homology. They viewed cladistics as agnostic about history, and that ‘evolutionary histories’ based on cladograms were nothing more than futile speculation. Pattern cladists Ebach et al. summarize it like this:
“Cladistics is not about evolution, but about the pattern of character distribution in organisms, or the recognition and characterization of groups.”7
This argument spilled over into the ‘creation science’ controversies of the day.8 Some candid statements of Colin Patterson, a noted critic of Hennigian cladistics, were particularly influential in the controversy.9 He discounted the speculative evolutionary reconstructions many systematists attached to their cladistics analyses because there was no way to identify in reality the putative ancestors ‘identified’ by the nodes on a cladogram:
“As the theory of cladistics has developed, it has been realized that more and more of the evolutionary framework is inessential, and may be dropped. The chief symptom of this change is the significance attached to nodes in cladistics. In Hennig’s book, as in all early works in cladistics, the nodes are taken to represent ancestral species. This assumption has been found to be unnecessary, even misleading, and may be dropped.”10
This raised a rather pertinent question: why invoke evolution at all if there’s no way to reconstruct evolutionary history from morphological comparisons? It’s very well to acknowledge that one believes in evolution, but transformed cladists essentially threw out the fossil record as evidence for it. With no fossil record and no viable biological mechanism for evolution, transformed cladists were left with a thoroughly unscientific ‘evolution-of-the-gaps’ mentality. It’s not surprising Hennigian cladists didn’t like it; transformed cladistics validated the creationist critique of evolutionary ‘reconstructions’ from the fossils!
Acceptance
With the availability and power of computers in the 1990s, cladistics became much easier to do, since before then weighing up cladograms with more than about 15 characters included in the analysis was unwieldy.11 Much of the contention then died down and Hennig was essentially proclaimed the victor.9,12 Hennigian cladistics ended up becoming the dominant cladistics method used by systematists today, more on the strength of academia’s commitment to evolution than the actual dependence of the cladistics method on evolution. However, many of the important issues raised in the 1970s and 1980s remain contentious today, though they are rarely talked about as openly.
Incompatibility of cladistic assumptions and evolution
Despite the fact that cladistics was originally intended to demonstrate evolution and the most probable phylogenies, there are a number of assumptions essential to cladistics methodology that make it ill-suited to demonstrating evolution.
Defining discrete, independent variables in biology
Cladistics regards all characters within the analysis as discrete, independent variables. However, biology can hardly be described in solely discrete terms; there are many features of animals that are continuous. Moreover, there are complex interdependencies within biology from the molecular to the organismal level, many of which we don’t currently understand. Therefore, defining a character for cladistics analysis even in genetics can be incredibly difficult. This problem generally becomes more pronounced with complex morphological features such as bones, as Lieberman points out:
“Bones have generally low degrees of heritability because they form parts of complex, integrated functional units that are subject not only to many genes with multiple effects (pleiotropism), but also to a large number of nongenetic influences. It is therefore difficult to divide bones into discrete, independent units of phylogenetic information. For these reasons, bones and other aspects of morphology can yield reasonably correct results for phylogenetic analyses of high-level taxonomic units, but become increasingly less reliable at lower taxonomic levels, such as species.”13
One of the major reasons for this problem is scale—the smaller one defines the morphological characters used for analysis, the larger the problem of interdependence generally becomes. Therefore, character selection becomes less reliable, and so do any interpretations of homology that are based on them. And since homology can really only exist in any meaningful way for evolution at the species level, it is practically impossible to demonstrate evolution using cladistics methodology unless one assumes evolution from the outset.
‘Shared derived characteristics’ and the illusion of lineages

Synapomorphies, or ‘shared derived characters’, are the hallmark of Hennigian cladistics.14 These are contrasted with symplesiomorphies, or ‘shared ancestral characters’, which cladists believe don’t possess any useful information for cladistics analysis (figure 2). The difference between the two is formally indistinguishable unless a character rooting procedure is used, which gives direction to the cladogram.15
In cladistics, synapomorphies are usually equated with homologous characters, which are understood as evidence of common ancestry.11,16 As noted above, there are difficulties in defining ‘character’ in biology in useful ways for cladistics analysis. However, when we take these limitations into account, we can still arrive at a fairly accurate description of the morphological patterns of similarity throughout multicellular life through a comparison of ‘shared’ characters.
The major problem with shared derived characters, however, rests with the word ‘derived’. Similarity in form does not guarantee a common ancestry, and this is an interpretation of the cladogram. Patterson pointed out quite aptly:
“Yet Gould and the American Museum people are hard to contradict when they say there are no transitional fossils. … I will lay it on the line—there is not one such fossil for which one could make a watertight argument. The reason is that statements about ancestry and descent are not applicable in the fossil record. Is Archaeopteryx the ancestor of all birds? Perhaps yes, perhaps no: there is no way of answering the question. It is easy enough to make up stories of how one form gave rise to another, and to find reasons why the stages should be favoured by natural selection. But such stories are not part of science, for there is no way of putting them to the test.”17
Note the biggest problem: there is no way to tell from the fossils if any lineage has been preserved. Cladistics doesn’t demonstrate evolution. Cladistics is a classification scheme, nothing more. Woodmorappe illustrates this point very well (figure 3):
“But doesn’t the fact that organisms lend themselves to being arranged in nested hierarchies of polarized traits (that is, cladograms) itself prove that they evolved that way (or at all)? Hardly. Assuming evolution a priori, one could construct a cladogram that has an 18-wheel truck as its crown group, and which shows a clearly transition-filled, incremental appearance of ‘truckness’, beginning with the stem-group unicycle. Note also that the human, elephant, and bat is each highly-derived fish, just as an 18-wheel truck is a highly-derived unicycle. Such is the reductio ad absurdum of cladistic methodology.”18
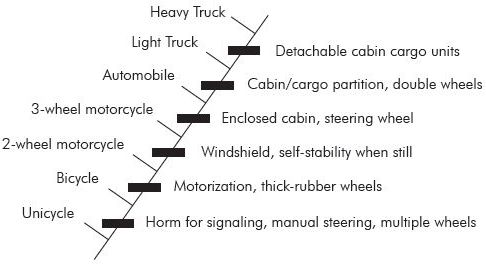
There is no reason to assume that the nodes of a cladogram represent putative ancestors, and there are no direct lineages demonstrated.19
What is a clade?
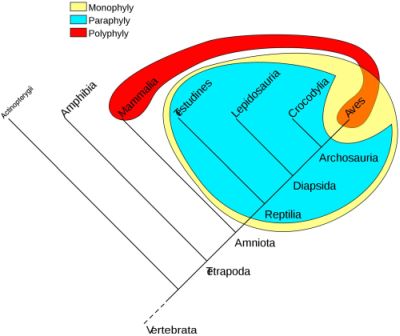
Typically, a clade is defined by ancestor-descendant relationships; it is an ancestral organism and all its descendants. All taxa within a clade are said to be monophyletic, i.e. they contain all the descendants of the possibly hypothetical closest common ancestor of the members of the group. Cladistics, by discovering synapomorphic characters, is meant to be able to distinguish monophyly from paraphyly (figure 4). A paraphyletic group is a monophyletic group minus one clade within that monophyletic group. An example is reptiles, a group which doesn’t include birds in common parlance, because evolutionists believe birds are supposedly descended from reptiles (theropod dinosaurs). Endothermic animals (mammals and birds) would be an example of a polyphletic group because endothermy is a homoplastic character, according to evolution.
However, we again run into the problem that cladistics methodology does not demonstrate a lineage. The ancestor is not just unknown in paleontology; it is unknowable. But the evolutionary extent of a clade is also arbitrary: it could be anything from a single parent and daughter to all biology (since orthodox evolution postulates everything arising from one common ancestor, not many). Cladists contend that all other methods of classification are biologically useless because genealogy is the only means of demonstrating the relative similarity between different organisms. However, Ernst Mayr points out that cladists don’t fully appreciate the genetic and phenotypic distance that may exist between supposed ‘sister taxa’:
“One of several phyletic sister lines may enter a new adaptive zone and there become exposed to severe novel selection pressures. As a result it will diverge dramatically from its cladistically nearest relatives and may become genetically so different that it would be biologically misleading to continue calling the sister groups near relatives. Yet being the joint descendants of a stem species they must be designated sister groups. And being sister groups they must be coordinate in rank, i.e., according to cladistic theory, they must have the same absolute categorical rank in the hierarchy (Hennig 1966, 139). This decision ignores the fact that one is still very much like the stem species while the other has evolved in the meantime into a drastically different type of organism.”20
Mayr also makes a distinction between different types of divergence that cladistics fails to identify: “Any theory of classification which pays no attention to the tremendous range of difference between shifts of phyletic lines into minor niches and into entirely new adaptive zones, is bound to produce classifications that are unbalanced and meaningless. But such a neglect of different kinds of phyletic evolution is precisely what the cladistic method demands.”21
Cladistics only acknowledges changes that arise due to a branching pattern (cladogenesis), and thus homogenizes different types of morphological disparity, producing a biologically unrealistic situation. Ironically, this makes phyletic gradualists (such as Mayr) unwitting allies with creationists because we too acknowledge different types of morphological disparity. The difference of course is that (in spite of the evidence) Mayr et al. are convinced that large-scale morphological discontinuity can be bridged naturalistically—creationists are not. Nevertheless, this inability to differentiate between different types of biological disparity renders cladistics inadequate for reconstructing the history of biology.
Evolution does not require a nested pattern
Cladistics assumes that its units for comparison can be arranged in a nested hierarchy.22 Evolutionists assume that evolution is the only viable explanation for a pattern of nested hierarchy. Hennigians go a step further, and then say that that makes evolution a viable process theory which gives cladistics real-world meaning, justifying its use in systematics.23 However, this is demonstrably untrue. Patterns of nested hierarchy in nature are not dependent on evolutionary assumptions since they were recognized well before naturalistic evolution was accepted by the scientific community:
“Although it is not in principle demonstrable from external evidence (Panchen, 1992), the existence of a single, irregularly branching hierarchy of relationships among biological taxa has been considered an empirical fact by Brady (1985), based on its historical emergence as the predominant means to represent patterns of taxonomic grouping used by pre-evolutionary systematists during the early 19th century. That this occurred prior to the general acceptance of evolutionary theory by the scientific community is clear evidence that a hierarchical conception of the Natural System is not dependent on an evolutionary process theory (Crow, 1926; Platnick, 1982).”24
If evolution was not required to conceive of life as a nested pattern, then life’s nested pattern is accommodated by evolution, not predicted or verified by it. When Hennig tries to establish the theoretical priority of evolution on nested hierarchy,23 he fails to see his anachronistic and ill-founded assumption of naturalism. Darwin assumed the nested pattern of life that had already been demonstrated independently of evolution. He then constructed an explicitly naturalistic explanation for its origin.
However, evolution does not demand a nested pattern because it can accommodate other patterns just as easily, if not more so.25 For instance, transposition (also known as lateral gene transfer) would provide a much faster mechanism than common descent for disseminating new genes/structures throughout the biosphere. Evolutionists would still assume descent with modification occurred because it provides the mechanism for biological novelty. But widespread transposition would add so much noise to any nested pattern assumed to be congruous with descent with modification that the nested pattern would be lost. Evolutionists don’t accept transposition as a widespread phenomenon, especially in multicellular life, simply because patterns that suggest transposition are not observed.
Moreover, not even common descent requires a nested pattern.26 Since characters are assumed to have independent phyletic histories and rates of evolution, there is no guarantee that even close sister taxa will have relatively similar morphology in comparison to more distantly related organisms. Moreover, transformation within a lineage (anagenesis) does not produce a nested pattern because the transformation that supposedly occurred was not caused by a branching event. Homoplasy confuses the issue even further because it can make distantly related creatures more morphologically similar than supposed sister taxa. Common descent has access to a veritable grab-bag of explanations that need not produce a nested pattern.
Pattern cladists, though they dismiss evolution as theoretical justification for cladistics, still believe it is the only viable explanation for it. However, common design also explains such a pattern, and with potentially more force.27 If life is designed to send a robust message that it is the product of one designer, nested hierarchy does the job. Even if the message receiver (us) has vastly incomplete comprehension of the data (through species extinction, or inability to investigate all the data), a nested pattern unifies life, is filled with homoplasies, and also presents large enough morphological gaps between different life forms to foil common descent. Life thus sends a unified non-naturalistic message: it is the product of one designer who designed life to resist naturalistic explanations for its origin.
Cladistics demands a nested pattern, and the fossil evidence fits into such a pattern relatively well, especially for higher taxonomic categories. However, neither evolution in general nor descent with modification in particular demand a nested pattern. Moreover, the nested pattern can be explained at least as well in a common design paradigm. Therefore evolution cannot claim to be the logical justification for cladistics, and it’s not the only available explanation for such a nested pattern. Neither can evolutionists legitimately consider cladistics an accurate reflection of actual phylogeny because evolution demands anagenesis, not just cladogenesis.
Problems in results and interpretation
Homology
The problems that the concept of homology presents for evolution in general have been well documented elsewhere, and will not be revisited here.28 However, there are a few important comments to make regarding homology and the cladistics method. Cladistics as a methodology may help identify homology, depending on the definition of homology that’s used. Defining homology with respect to cladistics analysis has proven as difficult as it has with respect to other systematic methods.16 The term ‘homology’ originated with Richard Owen, and he only saw it as similar structures used for different functions. Darwin defined homology in a similar way:29
“All physiologists admit that the swim bladder is homologous, or ‘ideally similar’, in position and structure with the lungs of the higher vertebrate animals … .”
The rest of the sentence shows he interpreted homology as providing support for common ancestry:
“ … hence there seems to me to be no great difficulty in believing that natural selection has actually converted a swim bladder into a lung, or organ used exclusively for respiration.”
It was only post-Darwinian biologists that defined homology as “similar structures resulting from common descent”. They defined a common designer explanation out of existence.
Some transformed cladists have recognized this distinction and have since abandoned the ‘traditional’ evolutionary definition of homology and adopted something closer to Owen’s descriptive definition.30 Homology thus may or may not demonstrate common descent, but common descent is irrelevant to the cladistics relationship because common descent becomes a historical explanation for homology rather than homology by definition.
Homoplasy
One of the biggest questions facing evolutionists regarding their morphological analyses is how to distinguish homoplasy from homology. Homoplastic traits are similar in function, but have different underlying structures, and as such cannot be explained by common descent. An obvious example is different types of wings: the wings of insects, birds, bats and pterosaurs all have very different structures, but have the same function—flight. Even the most ardent defenders of Hennigian cladistics take it for granted that homoplasy is common in cladistics analyses.31 Homoplasy creates noise in any cladogram because it can lead to false identifications of homology if not properly identified. However, the problem becomes one of scale—the smaller one subdivides characters to gain more characters, the more subjective character selection becomes. Woodmorappe points this out:
“ … whereas a nested hierarchy may well characterize living things when viewed in terms of general similarities and differences, it does not exist when large numbers of detailed morphological similarities and differences are simultaneously considered.”32
Moreover, since many structures that were assumed to be homologous at the morphological level have since been shown to be homoplastic at the molecular and/or developmental levels,28 the argument from homology is consistently getting weaker.
Mosaic evolution
A corollary for evolutionists in defining morphological traits as independent variables is that individual characters have independent phyletic histories, otherwise known as mosaic evolution. Individual traits can have evolutionary rates that speed up, slow down, stop, or reverse, all independently of other characters. However, if characters can evolve like this, why should we expect a nested pattern as opposed to any other? Such a concept can explain anything, which makes it unfalsifiable.
It also means that organisms that possess a combination of fully formed characters found in different clades are called ‘intermediate’ or ‘transitional’ fossils. However, such creatures are better termed ‘mosaics’, and they were fully functional. Moreover, no transformation has been demonstrated, only for example, fish with some tetrapod characters (Tiktaalik)33 or birds with some reptilian features (Archaeopteryx).34
Essentially, mosaic evolution is ubiquitous homoplasy without a discernible evolutionary pattern. Mosaic evolution thus has limitless explanatory scope; but it comes at the high price of sacrificing all explanatory power. Evolution needs an empirically demonstrable mechanism for historical plausibility, and no viable ones have ever been demonstrated, nor are they likely to be.35,36 However, mosaic evolution exacerbates this problem 100-fold. All the discontinuities and reversals mosaic evolution purports to explain have to be accounted for mechanistically. However, such a mechanistic explanation would be hopelessly complex and contradictory because it would have to explain every possible evolutionary scenario at the same time. Mosaic evolution is thus a smokescreen that hides the fact that a mechanistic explanation for the fossil pattern it describes would be hopelessly complex and contradictory, not to mention biologically unrealistic.
One then wonders: if this is the case, would evolutionists ever use such a ridiculous explanation for the fossil patterns? Most major vertebrate evolutionary series: such as the evolution of tetrapods,37 birds,38,39 mammals40 and whales,41 have been found to possess many discontinuities and reversals in individual character states. Daeschler et al., amid all the fanfare of the discovery of the now famous ‘transitional fossil’ Tiktaalik, describe the fish-to-tetrapod fossil series (including Tiktaalik) in this manner:
“Major elements of the tetrapod body plan originated as a succession of intermediate morphologies that evolved mosaically and in parallel among sarcopterygians closely related to tetrapods, allowing them to exploit diverse habitats in the Devonian [emphasis added].”42
They are forced to invoke mosaic evolution because of the numerous discontinuities and reversals present in the series. This pattern is present in the majority of the major vertebrate fossil ‘evolutionary transitions’ despite 150 years of looking for the myriad transitional forms Darwin predicted. If evolution has to rely so heavily on mosaic evolution to explain fossil patterns, evolution simply cannot explain the patterns in the fossil record.
Emphasis on morphology at the expense of timeline
Cladistics focuses primarily on morphology while working with its own idealized timeline governed by cladogenesis. This is nominally fine for comparing extant creatures since there is no separate timeline for comparison. However, it is troublesome for evolutionary paleontology because there are frequent conflicts with fossil dating and the idealized morphological ‘timeline’ produced by a cladogram. As a result, many morphological analyses end up producing the ‘grandfather paradox’, where organisms deemed ‘ancestral’ by the cladistics analysis are actually reported by evolutionists to be millions of years younger than the supposed descendants. A recent example is tetrapod tracks from Poland that were ‘dated’ 20 Ma older than Tiktaalik, the heavily promoted ‘transitional fossil’ between fish and tetrapods.43 These are also often termed ‘ghost lineages’, since these supposed ‘ancestral’ organisms leave no trace in the fossils where they’re expected to be.
Sometimes there is ‘only’ a few million years’ difference, which renders the fossils prone to being ‘redated’, since a few million years either way is generally geologically insignificant to long-agers. However, one area where this is a major problem is orthodox dino-to-bird speculation. Evolutionist Peter Dodson sums up the problem nicely:
“Personally, I continue to find it problematic that the most birdlike maniraptoran theropods are found 25 to 75 million years after the origin of birds … . Ghost lineages are frankly a contrived solution, a deus ex machina required by the cladistic method. Of course, it is admitted that late Cretaceous maniraptorans are not the actual ancestors of birds, only ‘sister taxa’. Are we being asked to believe that a group of highly derived, rapidly evolving maniraptorans in the Jurassic gave rise to birds, as manifested by Archaeopteryx, and then this highly progressive lineage then went into a state of evolutionary stasis and persisted unchanged in essential characters for millions of years? Or are actual ancestors far more basal in morphology and harder to classify? If the latter, then why insist that the problem is now solved?”44
With regard to dinosaur-to-bird evolution, the irony is that this problem is perhaps worst for the most basal dramaeosaurid currently known (which are said to be the closest dinosaurian relatives of birds), Mahakala omnogovae.45 The extant fossils for Mahakala are ‘dated’ at 80 Ma, but the split between dramaeosauridae and paraves supposedly occurred about 140 Ma.46 Moreover, there are many dramaeosaurs that fill in that chronological gap, but they are all ‘more advanced’ in their morphology than Mahakala. This is a ghost lineage 60 Ma in the making!
Cladograms and the illusion of evolution
ReMine identifies cladograms as one of the main culprits in giving the illusion of evolution.47 The evolutionary tree is a powerful image that has been one of the hallmarks of evolution’s public image, and cladistics plays on that very hallmark because cladograms look very much like traditional evolutionary trees. It has power because it purports to demonstrate a ‘lineage’, which the public automatically interprets as akin to a family tree (figure 1).48 Pattern cladists are often quick to point out that cladograms are not lineages in the strict sense, but that they purport to be best-guess models of the path evolution took. At the same time, Hennigian evolutionists are keen on repeating the mantra that cladistics “is the purest of all genealogical systems for classification, since it works only with closeness of common ancestry in time [emphasis added].”8
Textbooks on cladistics can be laden with genealogical terms, as if cladistics and genealogy are speaking about the same thing.49 Words such as ‘ancestral’, ‘derived’, ‘lineage’, ‘genealogy’, ‘primitive’, ‘advanced’, etc. are constantly used to depict the relationships between taxa determined by cladistics to the public. However, cladistics never identifies ancestors: it uses myriad other methods to represent what an ancestor may have been like.15 This creates confusion because it makes cladograms look as if they are equivalent demonstrations of genealogy as family histories. Therefore, the continued use of terms loaded with genealogical connotations in the public arena will always mislead the majority of the public, who know little of the intricacies of biological systematics. There seems to be only one viable solution to avoiding this confusion: avoid the use of such terms.
I believe, however, that this honesty would inevitably come at a high cost. If the public truly understood what cladists mean, evolution would likely lose much public credibility because it would become evident that they can’t demonstrate the sine qua non of Darwin’s theory: descent with modification.
Conclusions
Fossils are fickle. They are fragmentary, sparse and open to contrasting and contradictory interpretations. Moreover, cladograms based on morphology have often been shown to be completely at odds with embryological and molecular data. When cladistics and fossil analysis are then combined, it results in a hopelessly subjective game of evolutionary theorizing, and has no power to independently verify evolution. This subjectivity is worsened since cladistics analyses are often completely at odds with fossil dating. Ad hoc hypotheses are usually required to harmonize the timeline implicit in the cladogram with the accepted fossil timeline.
Cladistics, by making cladogenesis the sole method of character state change, ignores different types of biological disparity. It simply extrapolates known mechanisms of speciation and assumes that they can produce complex novelty, which both creationists and many evolutionists reject. Moreover, defining characters for cladistics analysis is tricky because interdependent characters can skew analyses, which becomes worse with higher resolution character selection. By essentially ‘digitizing’ taxa and linearizing biological disparity, cladistics produces a biologically unrealistic situation and speaks little to the truth or falsity of evolution or creation.
Nevertheless, cladograms are paraded as demonstrations of evolution, and yet it fails to identify ancestors and descendants. Language connoting ancestry and the usage of cladograms in presentations of phylogeny is often used to convey what systematists understand as mere topology. The ‘evolutionary tree’ has been a powerful metaphor used to demonstrate evolution for the past 150 years, and cladograms play on this image in the public consciousness, whether the experts intend them to or not. This confuses the public because they misunderstand what the cladograms actually demonstrate.
Cladistics enables us to gain a picture of the nested hierarchy of life, but the method itself tells us nothing about what produced that pattern. Since evolution wasn’t needed either to discover or understand the pattern, it’s not evidence for evolution. Therefore, creationists need not worry about what cladistics purports to show. Nevertheless, this also stresses the need for a proper systematic method that can demonstrate biological disparity. Until creationists and evolutionists learn to communicate using a form of systematics that can empirically identify biological disparity, we will continue talking past each other.
The irony is that cladistics was developed by evolutionists for evolutionists, and it still fails to demonstrate evolution, let alone biological reality. This suggests that the problem lies not so much with the method, but with the underlying theory it purports to demonstrate. The cladograms are models of the pattern of life, and as such have limitations. However, reading evolution, which is inescapably genealogical, into a method that explicitly shies away from notions of genealogy, makes evolution look like it’s running from reality.
References
- Hennig, W., Grundzügeeiner Theorie der phylogenetischen Systematik, Deutscher Zentralverlag, Berlin, 1950. Return to text.
- Mayr, E., Classification and phylogeny, American Zoologist 5:165–174, 1965. Return to text.
- Hennig, W., Phylogenetic Systematics, University of Illinois Press, Urbana, IL, 1966. Return to text.
- Hennig, W., Phylogenetic systematics, Annual Review of Entomology 10:97–116, 1965. Return to text.
- Ridley, M., Evolution and Classification: The Reformation of Cladism, Longman, London, 1986. Return to text.
- Nelson, N. and Platnick, N., Systematics and Biogeography: Cladistics and Vicariance, Columbia University Press, New York, 1981. Return to text.
- Williams, D.M. and Ebach, M.C., Foundations of Systematics and Biogeography, Springer Science+Business Media, New York, p. 107, 2008. Return to text.
- Gould, S.J., Darwinism defined: the difference between fact and theory, Discover 8:64–70, 1987. Return to text.
- Nelson, P.A., Colin Patterson revisits his famous question about evolution, Origins & Design 17(2), 1997; www.arn.org/docs/odesign/od171/colpat171.htm. Return to text.
- Patterson, C., Cladistics, The Biologist 27:234–240, 1980. Return to text.
- ReMine, W.J., The Biotic Message, St Paul Science, St Paul, MN, p. 268, 1993. Return to text.
- Wiley, E.O., Siegel-Causey, D., Brooks, D.L. and Funk, V.A., The Compleat Cladist: A Primer Of Phylogenetic Procedures, Special publication no. 19, Museum of Natural History, University of Kansas, Lawrence, KS, 1991. This primer on cladists only mentions transformed cladistics very briefly as a heterodox branch of cladistics and offers no discussion of its merits, only references to standard rebuttals. This shows that even as early as 1991 transformed cladistics was considered little more than a historical footnote by most cladists, albeit a recent one. Return to text.
- Lieberman, D.E., Homology and hominid phylogeny: problems and potential solutions, Evolutionary Anthropology 7:142–151, 1999. Return to text.
- Wiley et al., ref. 12, p. 1. Return to text.
- Bryant, H.N., Character polarity and the rooting of cladograms; in: Wagner, G.P. (Ed.), The Character Concept In Evolutionary Biology, Academic Press, NY, pp. 319–338, 2001. Return to text.
- dePinna, M.C., Concepts and tests of homology in the cladistic paradigm, Cladistics 7:367–394, 1991. Return to text.
- Sunderland, L., Darwin’s Enigma, Master Books, Green Forest, AR, pp. 101–102, 1998. Return to text.
- Woodmorappe, J., Evolutionary cladograms and malevolent, strawmen creationists: a review of Evolution: What the Fossils Say and Why it Matters by Donald R. Prothero, J. Creation 23(3):39–43, 2009. Return to text.
- ReMine, ref. 11, p. 273. Return to text.
- Mayr, E., Cladistic analysis or cladistic classification? Zeitschrift fűr Zoologische Systematik und Evolutionforschung 12:94–128, 1974; p. 102. Return to text.
- Mayr, ref. 20, pp. 105–106. Return to text.
- Bryant, ref. 15, p. 330. Return to text.
- Hennig, ref. 4, pp. 100–101. Return to text.
- Brower, A.V.Z., Evolution is not a necessary assumption of cladistics, Cladistics 16:143–154, 2000. Return to text.
- ReMine, ref. 11, p. 358. Return to text.
- ReMine, ref. 11, p. 343. Return to text.
- ReMine, ref. 11, pp. 344–368. Return to text.
- Bergman, J., Does homology provide evidence of evolutionary naturalism? J. Creation (formerly TJ) 15(1):26–33, 2001. Return to text.
- Darwin, C., On the Origin of Species, 1st ed., ch. 6: Difficulties on theory, 1859; www.talkorigins.org/faqs/origin/chapter6.html. Return to text.
- Williams and Ebach, ref. 7, p. ix. Return to text.
- Farris, J.S., The logical basis of phylogenetic analysis; in: Platnick, N.I. and Funk, V.A. (Eds.), Advances in Cladistics, Proceeding of the second meeting of the Willi Hennig Society, vol. 2, Columbia University Press, NY, pp. 7–36, 1983. Return to text.
- Woodmorappe, J., Eviscerating Eldredge: A review of The Triumph of Evolution and the Failure of Creationism by Niles Eldredge, J. Creation (formerly TJ) 15(2):13–16, 2001. Return to text.
- Sarfati, J., Tiktaalik—a fishy ‘missing link’, J. Creation 21(1):53–57, 2007. Return to text.
- Woodmorappe, J., Bird evolution: discontinuities and reversals, J. Creation 17(1):88–94, 2003. Return to text.
- Sanford, J., Genetic Entropy and the Mystery of the Genome, 3rd edn, FMS Publications, New York, 2008. Return to text.
- Williams, A., How life works, J. Creation 22(2):85–91, 2008. Return to text.
- Garner, P., The fossil record of ‘early’ tetrapods: evidence of a major evolutionary transition? J. Creation 17(2):111–117, 2003. See also Sarfati, J., Tiktaalik—a fishy ‘missing link’, J. Creation 21(1):53–57, 2007. Return to text.
- Woodmorappe, J., Bird evolution: discontinuities and reversals, J. Creation 17(1):88–94, 2003. Return to text.
- Oard, M.J., Did birds evolve from dinosaurs? J. Creation 25(2):22–31, 2011. Return to text.
- Woodmorappe, J., Mammal-like reptiles: major trait reversals and discontinuities, J. Creation (formerly TJ) 15(1):44–52, 2001. Return to text.
- Woodmorappe, J., Walking whales, nested hierarchies, and chimeras: do they exist? J. Creation (formerly TJ) 16(1):111–119, 2002. Return to text.
- Daeschler, E.B., Shubin, N.H. and Jenkins Jr, F.A., A Devonian tetrapod-like fish and the evolution of the tetrapod body plan, Nature 440(7085):757–763, 6 April 2006. Return to text.
- Walker, T., Tetrapods from Poland trample the Tiktaalik school of evolution, J. Creation 24(1):39–42, 2010. Return to text.
- Dodson, P., Response by Peter Dodson, American Paleontologist 9(4):13–14, 2001; cited in, Woodmorappe, ref. 38. Return to text.
- Doyle, S., Plucking the dinobird, 28 September 2007. Return to text.
- Turner, A.H., Pol, D., Clarke, J.A., Erickson, G.M. and Norell M.A., A basal dromaeosaurid and size evolution preceding avian flight, Science 317:1378–1381, 7 September 2007. Return to text.
- ReMine, ref. 11, pp. 277–301. Return to text.
- Silvestru, E., Flying dinosaurs, flightless dinosaurs and other evolutionary fantasies, J. Creation 20(2):42–47, 2006. Return to text.




Readers’ comments
Comments are automatically closed 14 days after publication.