Journal of Creation 16(1):15–25, April 2002
Browse our latest digital issue Subscribe
A baraminology tutorial with examples from the grasses (Poaceae)
Creationist biosystematics has existed since Frank Marsh coined the term baramin in 1941. Unfortunately, actual research into identifying baramins has been sparse. In the past decade, creation biologists have worked to develop a systematic methodology called baraminology. This paper presents a short tutorial on some of the techniques now in use to identify and study baramins. Readers are encouraged to use the information in this paper as a starting point for baraminology research of their own.
The biological discipline of systematics was developed to discover natural groupings of organisms, such as species. A new systematic method, baraminology, specifically pertains to creationists.1 Baraminology seeks not the species but the baramins, God’s ‘created kinds’. In the broadest sense, baraminology has its roots in the writings of Frank Marsh. In 1941, Marsh coined the term baramin.2 However, Marsh’s ideas have begun to flourish in creationist research only in the past two decades. The German creationist group Wort und Wissen has produced a book of systematics papers, Typen des Lebens, in which they apply Marsh’s ideas to groups of plants and animals.3 Fortunately for English-speaking creationists, Georg Huber is currently translating the book into English. Also during the 1990s, Kurt Wise applied baraminology to turtles,4 and Ashley Robinson and David Cavanaugh produced a series of papers on baraminology in turtles,5 primates6 and cats.7 I have been very active ‘behind the scenes’ in promoting baraminology to my fellow biologists. As part of the Baraminology Study Group (BSG), I helped organize two baraminology conferences at Liberty University and Cedarville University.8,9
Science in general and baraminology specifically require an appropriate philosophical basis in order to be successful in describing the world. At the baraminology conferences, so much emphasis has been placed on philosophy that researchers have not gained a practical understanding of the basic methodology and relevance of baraminology. Consequently, I find that many researchers do not know how to proceed. In this short work, I intend to demonstrate as clearly as possible how to undertake a baraminology study, using the grass family Poaceae as an example. It is my hope that once others see how straightforward it can be, they will be encouraged to try it themselves.
What to look for
Many creationists share the problematic desire to have a definition of baramin that makes it easy to recognize. Marsh’s heavy emphasis on hybridization as the defining feature of a baramin has certainly contributed to this bias.10 An unambiguous criterion makes research easy, but even the hybridization criterion has serious limitations (e.g. it is inapplicable to asexual or fossil organisms). Because of these problems, baraminologists of today focus on approximating the limits of the baramin using a suite of characteristics. To assist in the approximation, we employ three terms that are derived from Marsh’s baramin:11
- The monobaramin is a group of organisms that share continuity, either genetic or phenetic.
- The apobaramin is a group of organisms that is discontinuous with everything else. Creationists have long used bats as an example of animals that are unrelated to any other mammals.12,13 Since we don’t know how many kinds (baramins) of bats God created, baraminologists refer to the bats as an apobaramin.
- The holobaramin is roughly what we call the ‘Genesis kind’. Technically, it simply combines the definitions of monobaramin and apobaramin. A holobaramin contains a complete set of organisms that share continuity among themselves but are discontinuous with all other organisms.
Because these definitions are not mutually exclusive, they form the basis of the baraminological method of successive approximation. If you divide groups of organisms into smaller and smaller apobaramins by subtractive evidence, you will eventually come to a point when you can legitimately divide the group no longer. Similarly, if you add more and more species to a monobaramin by additive evidence, you will eventually come to a point when you cannot legitimately add any more species. Hopefully, the point at which the apobaramin can no longer be divided and the point at which the monobaramin can no longer be expanded is the same point: the holobaramin. At this point, the ‘membership list’ of the monobaramin and the apobaramin are exactly the same; therefore, this group probably represents the holobaramin.
To do baraminology then, we evaluate two kinds of evidence: Additive and subtractive. Hybridization works well as additive evidence. The ability of members of two different species to produce offspring strongly indicates that they share basic genetic machinery and a common developmental path; however, failure to hybridize is not subtractive evidence. There are too many factors that can cause reproductive isolation that have nothing to do with baraminic status. Unfortunately, subtractive evidence proves difficult to identify in many cases. Sometimes the creation record in Genesis can provide the strongest subtractive evidence. For example, we know that whales share no ancestry with land mammals (Gen. 1:20–21).
If subtractive evidence cannot be found, you should not consider your baraminology study a failure:
- You might be looking at only part of the holobaramin; that is, your focus is too narrow. Prior studies have shown that the holobaramin is larger than most genera.
- Baraminology constantly advances and refines its methodology. Discontinuity that is undetectable today may be detected tomorrow.
- Practically speaking, establishing a monobaramin is useful information. For example, in a baraminology study of a group of species in the sunflower family, I found good evidence for continuity (hybridization) but no discontinuity with other species of the same family.14 At the very least, my results indicated that the holobaramin is broader than this group.
The grasses: choosing a subject
Biologists reading this article probably have a research subject in mind, but for those who do not, guidance on choosing a group may be in order: First, realize that you will likely choose a group that no creationist has studied before. Because precious little baraminological research has been published, you will probably not choose one of the few groups that have already been studied. Studying a group that has been the subject of previous baraminological analysis is also good. The essence of the baraminology method is approximation, so follow-up studies are always welcome.
Also consider how your baraminology study might relate to others already published. Will you study a group similar to one already studied, or will you choose something completely new? For example, since the dogs,15 bears16 and cats7 have all been the subjects of baraminology studies, another carnivore group, such as the weasels or raccoons, would complement the previous work well. On the other hand, studying a new group (e.g. invertebrates, microbes, or fungi) will blaze new trails in baraminology and expand our understanding of the general features of the baramin.
Practical issues involved in gathering appropriate data for your group of interest should be considered as well. Will there be enough published data to do a good baraminology study, or are you willing and able to gather your own data? Re-interpreting published data is less laborious than gathering new data, but published datasets can be sparse. For example, I was surprised to find almost no published, family-level cladistic (tabulations of shared / non-shared characters) datasets on dinosaurs. On the other hand, baraminologists need to begin generating our own data rather than simply re-interpreting what someone else has already published. If you are able, I would strongly encourage collecting your own data.
Most importantly, consider the biblical constraints that will inform the interpretation of your results. Even if the Bible does not specifically mention your organisms, the outline of early history in Genesis 1–11 will impact all baraminology studies. At the minimum, try to determine on which day of Creation your group originated and how your group survived the Flood (if it did). These aspects will be important for understanding the historical development of the baramin.
To illustrate the baraminological method, I have chosen the grasses. The grass family Poaceae is one of the most important families on the planet. People associate the word ‘grass’ with the stuff in their lawns, but grasses also include important cereal crops such as rice, maize, oats, wheat, barley, rye, and sugarcane. Half of the world’s population subsists on members of the grass family. The family itself consists of approximately 10,000 species in 5–6 subfamilies and 46 tribes.17
In addition to its utilitarian importance, Poaceae makes an excellent baraminology subject for a number of other reasons. First, a number of grasses are mentioned in the Bible, including barley (Ruth 2:23, Hosea 3:2), millet (Ezekiel 4:9, 27:17), wheat (Genesis 41:22, Leviticus 23:14), and the comprehensive term grass (Genesis 1:11–12). Second, because of the importance of the grasses, many botanists actively research Poaceae systematics. Scientists have formed a collaborative group to study the phylogeny of the grasses, and several genomics projects are underway for the more important cereal crops, mainly rice18 and maize.19 A great deal of data from these research projects is publicly available. Third, a creationist study of the wheat tribe has been published in Typen des Lebens,20 allowing a comparison of results and conclusions. Finally, my own research work has focused on rice, so grass baraminology will help me understand other areas of my research interests.18,21
The baraminology method
There really is no single ‘baraminology method’ but rather a collection of methods used in successive approximation. In the following sections, I present a few techniques that can be used by nearly any biologist. I begin with Scriptural considerations, then move to additive and subtractive evidences, and conclude with an interpretation of my results. At each step, I present general methods that can be applied to any group and illustrate their application in my study of the grasses. This paper is necessarily short, so some methods in baraminology have been omitted. Consult the literature for discussions of phylogenetic discontinuity detection,4 the use of mitochondrial DNA,5 and Analysis of Pattern.14,22
Biblical considerations
Because the Bible is the only source for infallible information, studying biblical passages greatly aids the identification and interpretation of baramins. The creation account can give clues about apobaraminic limits, and early references in Genesis and Job can illuminate the tempo and mode of post-Flood diversification (Job was originally written during the time of Abraham, approximately 500 years after the Flood). Unfortunately, many groups are not mentioned in the Bible, and others are mentioned in passages that are difficult to interpret. In these cases, little biblical evidence can be cited outside of the general outline of history in Gen. 1–11.
When a species or group of species is mentioned in the Bible, proper interpretation becomes very important for applying the passages to baraminology. Optimally, trained, careful Hebrew and Greek exegesis should be performed on the relevant texts by appropriate scholars. Since scholarly exegesis may be difficult to obtain, we can still benefit from our own preliminary study, with the recognition that we may be wrong. For the lay Bible student, variety of sources is the key to locating and understanding relevant biblical texts. Relying on one translation or commentary may lead to an enigmatic or peculiar understanding of a passage. Using a variety of translations and other resources will ensure that a balanced view of the passage is achieved. Although Scripture should not be interpreted by majority rule, alternative translations can alert the careful student to potentially valid alternative interpretations.
To begin a biblical study, list words that refer to your group and which might be found in English translations. For the grasses, this list includes most of the cereal crops: wheat, barley, etc. Next, use a concordance such as Strong’s Exhaustive Concordance or Young’s Analytical Concordance to locate specific verses that contain these words. Alternatively, the Bible Gateway (bible.gospelcom .net/bible) offers word searching in many different translations in fifteen languages, including the King James Version and the Latin Vulgate. I found that the Bible refers to members of the grass family frequently. I will focus my discussion on two types of passages: the creation of grasses and early post-Flood references.
The English translation of relevant passages should be verified by comparing translations and consulting lexicons and commentaries. I found eleven Hebrew words in Strong’s that are used in various passages to refer to the grasses. Using the Bible Gateway, I constructed a chart of the translations of these words from sixteen verses in five different translations (KJV, NKJV, NASB, RSV, NIV). On Bible Gateway web pages, different translations of the same verse can be viewed with the click of a mouse, greatly simplifying this analysis. Based on my chart (Table 1), I infer two important points. (1) The Hebrew word deše’ in Gen. 1:11–12 is translated ‘grass’ in the KJV and NKJV but is translated ‘vegetation’ in the NASB, RSV, and NIV. The variation in translation alerts me to possible scholarly disagreement over the meaning of the verses that record the creation of grass. (2) I also note on the chart that eight of the eleven words listed are found in Job. Two of these words, `ēśeb and hāsîr, are translated ‘grass’ in all five translations. The remaining six are agricultural words. Some (hittâh and śe`ōrāh) refer to crop species, while others refer to aspects of crops related to farming (e.g. sheaves, heads of grain, fodder, etc.).

Click here for larger view
I turned to additional resources to verify my understanding of these translation differences. First, I consulted the New International Dictionary of Old Testament Theology and Exegesis (NIDOTTE), edited by W.A. VanGemeren. This five-volume dictionary of Hebrew words has a helpful index in volume five that relates the words in the dictionary to the numbering system in Strong’s. NIDOTTE should be available in seminary libraries, or it can be purchased for around US$100. The dictionary entries on the words in Table 1 confirmed my interpretation from comparing translations.
Commentaries disagree over the interpretation of deše’ in Genesis 1:11–12. Some scholars believe that deše’ is a general descriptor for all vegetation, of which ‘herbs’ and ‘trees’ are the two main classes. Others maintain that there are three classes of plants, ‘grass’, ‘herbs’ and ‘trees’. The majority favour the first view.23-25
From this brief biblical survey, we may draw a few preliminary conclusions. First, the creation account in Genesis 1:11–12 does not directly address the origin of the Poaceae. In fact, the term deše’ is most frequently used for the green growth that sprouts in response to rain.26 An apobaraminic division between herbaceous plants and woody trees is also not required. God most likely created many individual plants to cover the newly formed land, including many members of the same baramin. If baramins were created with original diversity, woody and herbaceous plants could be members of the same baramin. Because modern plant baramins contain both woody and herbaceous members (e.g. Flaveriinae14 ), it is best to refrain from asserting one interpretation over another. I conclude that the creation account gives very little information about the baraminic limits of the grasses with respect to other plants.
The numerous agricultural words found in the book of Job form the basis of my second conclusion. The various farming terms indicate that an advanced agriculture already existed at the time of Job. Job speaks of barley (śe`ōrāh) and wheat (hittâh) using Hebrew words that refer unequivocally to these species.27,28 Since barley and wheat interbreed29 (placing them in the same monobaramin), their early cultivation indicates either a rapid post-Flood diversification of the baramin or a pre-Flood diversification preserved via seeds through the Flood. Since we know that Noah preserved food on the Ark (Gen. 6:21), pre-Flood domestication of wheat and barley could be a valid interpretation.
Additive evidence: hybridization
Due to its popularity, I will present hybridization as the first scientific method. If you are working with a group that is not amenable to hybridization experiments, you might want to skip to the next section on Robinson and Cavanaugh’s baraminic distance method, which can be used on any group.6 Space does not permit a full discussion of the theory of the hybridization criterion, so I recommend consulting other references1,30 for more information.
Unfortunately, good compilations of hybridization records are difficult to obtain. The Center for Origins Research and Education at Bryan College is developing a computerized database of hybrids to assist in baraminology studies.31 Though the HybriDatabase (HDB) (www.bryancore.org/hdb) currently contains 2,711 hybrid records, I have gained valuable experience during the development of the HDB. I formulated an effective method of locating hybrid records.
First, consult the HDB. Although incomplete, it contains valuable information. For each hybrid, a complete literature citation is available at the click of a mouse. Second, try computerized search engines. PubMed (www.ncbi.nlm.nih.gov) offers free searching of mostly biomedical and molecular biology journals. Ovid (www.ovid.com) and Biosis (www.biosis.org) offer database searching of a wider array of biology literature for a subscription fee. Many public university libraries provide Ovid or Biosis searching to their patrons. Third, consult published hybrid compilations. Excellent sources include Gray’s Bird Hybrids32 and Mammalian Hybrids,33 the periodicals Plant Breeding Abstracts and Animal Breeding Abstracts, and numerous specialty compilations (e.g. Orchid Hybrids34). You may consult online university library catalogues or Bookfinder (www.bookfinder.com) to locate hybridization compilations. I recommend the two Breeding Abstract periodicals as comprehensive sources of papers on hybrids. Creationists often recommend Gray’s books,30 but some of the hybrids listed are not accepted as valid.35 In all cases, try to locate the original paper to confirm the hybrid success. Finally, if you find a research article on a hybrid of interest, scan the references for other hybrid records.
I found a plethora of grass hybridization information in Knobloch’s A Check List of Crosses in the Gramineae,29 Омдаленная Гибридиза Растений (Omdalennaya Gibridiza Rasteniĭ, The Remote Hybridization of Plants, a Russian book on distant plant hybridization),36 Watson and Dallwitz’s Grass Genera of the World17 and several papers in Plant Breeding Abstracts. I also used the AltaVista search engine (www.altavista.com) to locate other records of newer hybrids.37-40
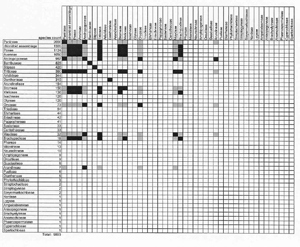
Click here for larger view
To display hybridization information, baraminologists frequently use a graphical tool called a hybridogram. To create a hybridogram, begin with graph paper or a computer spreadsheet. Next, list your species down the left side and across the top, forming a square matrix where each cell represents a potential interspecific hybrid (Figure 1). Record successful hybridizations by filling in the appropriate cells. The Wort und Wissen creationist group uses the hybridogram extensively in their book Typen des Lebens.3
The 10,000 grass species make a challenging subject for a hybridogram. Because I cannot put all species on one hybridogram, I made several approximations for the hybridogram in Figure 1. I listed only the 46 grass tribes recognized by Watson and Dallwitz.17 Next, I filled in cells indicating successful intergeneric hybridization within and between tribes. I also used Scherer’s secondary membership criterion, ‘Two individuals belong to the same basic type if they have hybridized with the same third organism.’30 By extension, I shaded cells grey where two tribes are known to cross with members of the same third tribe.
In Figure 1, inter-tribal grass hybrids join only twelve of 46 tribes. At first glance, 12 out of 46 seems like poor baraminic evidence, but the 12 hybridizing tribes comprise approximately 7,220 species. Consequently, I can assign 72% of the Poaceae to one hybridization-defined monobaramin. The remaining tribes that are not connected to the rest by hybridization are mostly small (half of the grass tribes contain less than 20 species). In his analysis of the duck baramin, Scherer noted the same pattern. Of the 13 tribes of the duck family Anatidae, hybridization connects eight. The remaining five represent tribes of 1–3 species each. Despite a lack of hybridization to connect the five small tribes with the remaining eight, Scherer still concludes that all Anatids (ducks, swans and geese) form a single basic type (or monobaramin; see below).41
Even though most of the non-hybridizing grass tribes are small, two tribes—Bambuseae (the bamboos) and Stipeae (including ricegrasses)—are quite large. This illustrates a limitation of hybridization: Lack of recorded hybridization is ambiguous baraminic evidence. Although I could find no hybrids between bamboos or ricegrasses and other grass tribes, my search for grass hybrids was cursory. A more comprehensive search may reveal hybrids that join all grass tribes. At this stage, I would advance the conservative hypothesis that 72% of grass species in 12 tribes form a monobaramin.
Additive and subtractive evidence: baraminic distance
Since hybridization is only additive evidence, I need more data to determine the apobaraminic status of Poaceae. Fortunately, Robinson and Cavanaugh developed statistical methods for examining baraminic relationships without hybridization data.6 They base their methods on the baraminic distance, a metric that summarizes systematic data. The information in systematic data sets is organized in columns where each column represents a particular characteristic, such as tooth shape or head size. The rows represent the taxa and the particular character states of those taxa. For example, oat flowers (character) are bisexual (character state 1) while maize flowers are unisexual (character state 2). For convenience, character states are almost always coded numerically (1=bisexual, 2=unisexual).
Systematic data sets can be challenging to locate. Systematists are aware of this limitation and have begun to archive their datasets in internet databases. You can use two different databases to search for datasets for your group of interest, TreeBASE (www.herbaria.harvard.edu/treebase/index.html) and Cladestore (palaeo.gly.bris.ac.uk/cladestore/default.html). Since the databases are relatively new, they only have a few datasets. You may need to dig further to find a useful dataset for your group. Specialty journals like Cladistics, Systematic Biology, and organism-themed publications (like Herpetologica or Journal of Mammalogy) often publish data sets to accompany articles on systematics. Although many published data sets exist, they are not always baraminologically useful. They may exclude taxa deemed baraminologically significant, or they may simply have too few taxa or characters to give reliable baraminic information. As mentioned previously, we creationists should strive to generate our own datasets by direct observations of living or preserved specimens. Only in this way can we obtain the precise data needed. In the meantime, published datasets can offer useful information in many cases.
Because of the importance of the grass family, the Grass Phylogeny Working Group (GPWG) placed a large data set online so that anyone with Internet access can analyze it (www.virtualherbarium.org/grass/gpwg/). The GPWG dataset contains 7,025 characters scored for 62 grass genera and four outgroup genera. The 62 grass genera represent 36 tribes. Most importantly, the large tribes excluded from the hybridization-defined monobaramin are present in this dataset; therefore, their baraminic status should be clearer. For more information about the GPWG dataset, consult their website.

Click here for larger view
Space prohibits a detailed explanation of the baraminic distance method, but a short description of the metric is in order. The baraminic distance between two species is the percentage of characters in which the two species differ in their character states. The simplicity of this metric is very important, because most evolutionary phylogenetic methods make assumptions of common ancestry to calculate similarities and distances. With a percentage, no prior assumptions are made, so identifying both significant similarity between species (implying baraminic relationship) and significant differences between other species (implying discontinuity) should be straightforward. For a detailed discussion of the baraminic distance method, consult Robinson and Cavanaugh’s original paper.6
I developed the computer program BDIST to perform the baraminic distance calculations on the large GPWG dataset. BDIST is available at the BSG website (www.bryancore.org/bsg), where you will also find detailed documentation on how to use the software. Because BDIST is written in Perl, it will run under any operating system. BDIST first sorts through the characters and calculates character relevance. Relevance is the percentage of taxa for which a character state is known, and BDIST includes relevance figures for each character in its output file. Robinson and Cavanaugh recommend that character with relevance less than 95% should be eliminated from baraminic distance calculations.6 After calculating relevances for every character, BDIST eliminates characters that have less than 95% relevance. Finally BDIST calculates baraminic distances from the remaining characters and outputs the distance matrix to a plain text file, which can be cut-and-pasted into a spreadsheet or other mathematical software for further analysis. BDIST eliminated 4,906 characters from the GPWG dataset because of low relevance. The remaining 2,119 characters were used for the baraminic distance calculations. Baraminic distances can be analyzed in a variety of ways. I will illustrate the correlation test, one application of baraminic distances.
Robinson and Cavanaugh recommend calculating the Pearson product-moment correlation between all possible pairs of taxa.6 If the distance between taxa A and B is similar to the distance between taxa C and B, and if this similarity of distances holds for taxa D, E, and F, then A and C are probably closely-related (Figure 2). By calculating the correlation of baraminic distances for taxa A and C, we can test whether the distances are similar enough to be statistically significant. Robinson and Cavanaugh suggest that significant positive correlation indicates that the two species are members of the same monobaramin and significant negative correlation indicates that the two species are discontinuous (members of different apobaramins). You should consult their paper for more information on baraminic distance correlation tests.6 I did not implement a correlation test in BDIST because these tests are more efficiently done by any number of statistical software packages. You can even use a simple spreadsheet, like Excel or QuattroPro. I use the S+ package, available from Insightful Corporation (www.insightful .com).

Click here for larger view
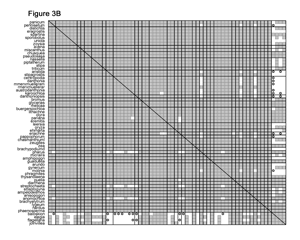
In the GPWG dataset, the 62 grass genera yield 1,891 unique species pairs for which baraminic distances and correlations can be calculated. Using the baraminic distances from BDIST, I found that 98% of the species pairs had significant positive correlation. Curiously, I also found that 53% of the 248 species pairs between the grasses and outgroup species also displayed significant positive correlation, and only 6% had significant negative correlation (Figure 3A). Based on Robinson and Cavanaugh’s original discussion of the distance correlation test, I did not expect a high frequency of significant positive correlation between the grass and outgroup species. These results suggest that the non-Poaceae genera included in the dataset might also be members of a monobaramin together with the grasses. If correct, this result would be very surprising, since grasses are widely acknowledged to form a well-defined group.
To re-evaluate these results, I removed molecular characters from the GPWG dataset and re-calculated the baraminic distances. Systematic data derived from DNA sequence comparisons may not be very useful for baraminology because so many DNA/DNA comparisons are done on genes that are very similar between many species. Consequently, species appear much more similar than they would if you examined their morphology, thus the use of DNA sequence information biases the systematic results towards similarity that is purely genetic.
Of the 7,025 characters in the GPWG dataset, only 53 are morphological. The remaining 6,972 characters come from DNA analyses. After eliminating the DNA characters, the baraminic distance calculations were very different. With the morphology-only dataset, 21 characters were eliminated due to low relevance, and 32 characters were used to calculate baraminic distance. From the Pearson correlation analysis, I found that nearly every one of the grasses shares significant positive correlation with all the other grasses but significant negative correlation with the outgroup genera. Two notable exceptions are the grass genera Streptochaeta and Anomochloa (possibly Pharus as well), both of which have significant negative correlation with most other grasses but significant positive correlation with the four outgroup genera and with each other (Figure 3B).
From the morphological analysis, I draw several conclusions. First, the Poaceae (excluding tribes Streptochaeteae and Anomochloeae) form a coherent monobaramin and apobaramin, suggesting that the majority of grass species are members of a single holobaramin. Second, negative baraminic distance correlation indicates that tribes Anomochloeae (1 sp.) and Streptochaeteae (2 spp.) are not members of the grass holobaramin. The position of Pharus and the Phareae (14 spp.) is presently unclear. Third and perhaps most important for the advancement of baraminology methods, heavy reliance on molecular sequence data biases baraminic analysis towards too much similarity. I strongly suggest that researchers do not rely too heavily on sequence similarity for determining baraminic relationships.
Conclusions
The final step of any baraminology paper is interpreting the analyses and presenting your conclusions. The considerations that went into selecting the group to study should now come back into play. You might consider the geographical distribution of the modern members of your baramin and how it relates to their Flood survival mode. You might also discuss possible diversification theories for an exceptionally large baramin. Relate your group back to the biblical references you already discovered and discuss their impact on both distribution and diversification. Finally, compare your results with the results of other creationist researchers. If you are dealing with a completely new group, discuss the general characteristics of your baramin, such as the number of species, the fossil record or how it compares with conventional taxonomic catagories (such as family, order or tribe).
Interpreting the grass holobaramin is a monumental task, so I will limit my comments to a few points. Junker previously assigned basic type status to the tribe Triticeae.20 Because basic type biology considers only hybridization and lacks a method of identifying discontinuities, a basic type is a monobaramin. Junker found no records of hybridization between species in the Triticeae and other tribes of the grasses. Since I found several intertribal hybridization records involving the Triticeae using the journal Plant Breeding Abstracts, I would broaden Junker’s basic type to include all the grasses except Anomochloeae and Streptochaeteae. In a report on the grass species Ring Muhly, the authors speculate that the boundaries of the ‘created kind’ lie within the genus Muhlenbergia.42 My results demonstrate that the holobaraminic boundaries of the grasses (including Ring Muhly) are much broader than any single genus.
Lastly, I want to address the question of the diversification of the grass holobaramin, the largest holobaramin identified to date. With 10,000 species, the grass holobaramin easily outnumbers even the biggest mammalian baramins. For example, a recent study places 150 fossil horse species into a single monobaramin.22 The great number of grass species is unlikely to be caused by excessive ‘splitting’ by over-zealous systematists. Instead, the large number of tribes indicates that the diversity is real. The fact that grasses are plants gives a possible clue to the origin of the extreme diversity. Unlike terrestrial animal baramins, many plant baramins survived the Flood with more than two individuals per baramin via debris rafting or preservation as food on the Ark. It is therefore possible that some of the grass diversity dates from before the Flood, possibly even from created diversity on Day 3 of the Creation Week.
Pre-Flood grass diversification would help to make sense of the early grass references in the Bible, particularly the advanced agriculture of Job. The species mentioned could have been preserved as food on the Ark. Some cereal grains might have arisen after the Flood. Archaeological evidence of a post-Flood domestication of barley (Hordeum vulgare) could be interpreted as merely diversification within the Hordeum genus.43 To clarify the issue of grass diversification, we will need to evaluate the post-Flood fossil record of the grasses.
With the Internet and the BDIST software, nearly any student or professional in biology can do a baraminological analysis of their favorite creatures. As we accumulate more baraminological studies, we will get a clearer picture of what baramins look like and how to identify them better. I pray that this article will help researchers become more familiar with baraminology and that biologists reading this article will seriously consider joining this exciting work.
Acknowledgements
I would like to thank David Fouts for advice on the biblical section of this paper and Stephanie Mace and Bevin Sims for critical readings of earlier drafts.
References
- Wise, K.P., Baraminology: A young-earth creation biosystematic method; in: Walsh, R.E. and Brooks, C.L. (Eds), Proceedings of the Second International Conference on Creationism, Creation Science Fellowship, Pittsburgh, pp. 345–358, 1990. Return to Text.
- Marsh, F.L., Fundamental Biology, Published by the author, Lincoln, 1941. Return to Text.
- Scherer, S. (Ed.), Typen des Lebens, Pascal-Verlag, Berlin, 1993. Return to Text.
- Wise, K.P., Practical baraminology, Journal of Creation 6(2):122–137, 1992. Return to Text.
- Robinson, D.A., A mitochondrial DNA analysis of the Testudine apobaramin, CRSQ 33(4):262–272, 1997. Return to Text.
- Robinson, D.A. and Cavanaugh, D.P., A quantative approach to baraminology with examples from Catarrhine primates, CRSQ 34(4):196–208, 1998. Return to Text.
- Robinson, D.A. and Cavanaugh, D.P., Evidence for a holobaraminic origin of the cats, CRSQ 35(1):2–14, 1998. Return to Text.
- Robinson, D.A. (Ed.), Baraminology ’99, Baraminology Study Group, 1999. Return to Text.
- Helder, M. (Ed.), Discontinuity: Understanding Biology in the Light of Creation, Baraminology Study Group, 2001. Return to Text.
- Marsh, F.L., Evolution, Creation, and Science, Review and Herald Publishing Association, Washington, 1947. Return to Text.
- ReMine, W.J., Discontinuity systematics: a new methodology of biosystematics relevant to the creation model; in: Walsh, R.E. and Brooks, C.L. (Eds), Proceedings of the Second International Conference on Creationism, Creation Science Fellowship, Pittsburgh, pp. 207–213, 1990. Return to Text.
- Nelson, B.C., After its Kind, Augsburg Publishing House, Minneapolis, 1927. Return to Text.
- Wysong, R.L., The Creation-Evolution Controversy, Inquiry Press, Midland, 1976. Return to Text.
- Wood, T.C. and Cavanaugh, D.P., A baraminological analysis of subtribe Flaveriinae (Asteraceae: Helenieae) and the origin of biological complexity, Origins, in press, 2001. Return to Text.
- Crompton, N.E.A., A review of selected features of the family Canidae with reference to its fundamental taxonomic status; in: Scherer, S. (Ed.), Typen des Lebens, Pascal-Verlag, Berlin, pp. 217–224, 1993. Return to Text.
- Tyler, D.J., Adaptations within the bear family: a contribution to the debate about the limits of variation, Creation Matters 2(5):1–4, 1997. Return to Text.
- Watson, L. and Dallwitz, M.J., Grass Genera of the World: <biodiversity.uno.edu/delta>, 23 Oct. 2001. Return to Text.
- Presting, G.G., Budiman, M.A., Wood, T., Yu, Y., Kim, H.R. et al., A framework for sequencing the rice genome, Novartis Found Symp 236:13–24, 2001. Return to Text.
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R. and Martienssen, R.A., Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome, Nat Genet 23(3):305–308, 1999. Return to Text.
- Junker, R., Der Grundtyp der Weizenartigen (Poaceae, Tribus Triticeae); in: Scherer, S. (Ed.), Typen des Lebens, Pascal-Verlag, Berlin, pp. 75–93, 1993. Return to Text.
- Mao, L., Wood, T.C., Yu, Y., Budiman, M.A., Tomkins, J. and others, Rice transposable elements: a survey of 73,000 sequence-tagged-connectors, Genome Res 10(7):982–990, 2000. Return to Text.
- Wood, T.C., Wise, K.P. and Cavanaugh, D.P., Pattern recognition analysis of fossil horses confirms the reality of the stratomorphic series; in: Helder, M. (Ed.), Discontinuity: Understanding Biology in the Light of Creation, Baraminology Study Group, p. 34, 2001. Return to Text.
- Paradise, B., Food for thought: the Septuagint translation of Genesis 1.11–12; in: Martin, J.D. and Davies, P.R. (Eds), A Word in Season, JSOT Press, Sheffield, England, pp. 177–204, 1986. Return to Text.
- Hamilton, V.P., The Book of Genesis Chapters 1–17, Eerdmans Publishing, Grand Rapids, 1990. Return to Text.
- Sarna, N., The JPS Torah Commentary Genesis, Jewish Publication Society, New York, 1989. Return to Text.
- Futato, M.D., Become green; in: VanGemeren, W.A. (Ed.), New International Dictionary of Old Testament Theology and Exegesis, Zondervan, Grand Rapids, pp. 999–1000, 1997. Return to Text.
- Wegner, P.D., Barley; in: VanGemeren, W.A. (Ed.), New International Dictionary of Old Testament Theology and Exegesis, Zondervan, Grand Rapids, pp. 1265–1266, 1997. Return to Text.
- Wegner, P.D., Wheat; in: VanGemeren, W.A. (Ed.), New International Dictionary of Old Testament Theology and Exegesis, Zondervan, Grand Rapids, pp. 104–105, 1997. Return to Text.
- Knobloch, I.W., A Check List of Crosses in the Gramineae, Published by the Author, East Lansing, 1968. Return to Text.
- Scherer, S., Basic Types of life; in: Scherer, S. (Ed.), Typen des Lebens, Pascal-Verlag, Berlin, pp. 11–30, 1993. Return to Text.
- Wood, T.C., Wise, K.P., Mace, S., Ingolfsland, K., Brown, M. and others, HybriDatabase: a computer repository of organismal hybridization data; in: Helder, M. (Ed.), Discontinuity: Understanding Biology in the Light of Creation, Baraminology Study Group, p. 30, 2001. Return to Text.
- Gray, A.P., Bird Hybrids: A Check-List with Bibliography, Commonwealth Agricultural Bureaux, Farnham Royal, Bucks, England, 1958. Return to Text.
- Gray, A.P., Mammalian Hybrids, Commonwealth Agricultural Bureau, Farnham Royal, Bucks, England, 1954. Return to Text.
- Sanders, F.H., Sanders’ Complete List of Orchid Hybrids, Gibbs & Bamforth Ltd., St. Alban, England, 1946. Return to Text.
- Friedmann, H., Georgia birds; bird hybrids; extinct and vanishing birds of the world, Science 129(3343):206, 1959. Return to Text.
- Цицин, Н.В., (Tsitsin, N.V.), Омдаленная Гибридиза Растений (Omdalennaya Gibridiza Rasteniĭ, The Remote Hybridization of Plants), Сельхозги (Sel’khozgi), Moscow, 1954. Return to Text.
- Jena, K.K., Development of intergeneric hybrid between O. sativa and Porteresia coarctata, Rice Genetics Newsletter 11:78–79, 1994. Return to Text.
- Sun, G., Yen, C. and Yang, J., Intermeiocyte connections and cytomixis in intergeneric hybrids III: Roegneria tsukushiensis X Psathyrostachys huashanica, Wheat Information Service 79:24–27, 1994. Return to Text.
- Sun, G. and Yen, C., The ineffectiveness of the ph1b gene on chromosome association in the F1 hybrid, Triticum aestivum X Psathyrostachys huashanica, Wheat Information Service 79:28–32, 1994. Return to Text.
- Yuan, W.-Y., Sun, S.-C., Liu, S.-X., Sun, Y., Tomita, M. and Yasumuro, Y., Production, fertility and cytology of tetrageneric hybrids involving Triticum, Agropyron, Haynaldia, and Secale, Wheat Information Service 84:13–18, 1997. Return to Text.
- Scherer, S., Der Grundtyp der Entinartigen (Anatidae, Anseriformes): Bioligische und Paläontologische Streiflichter; in: Scherer, S. (Ed.), Typen des Lebens, Pascal-Verlag, Berlin, pp. 131–158, 1993. Return to Text.
- Howe, G.F., Williams, E.L. and Meyer, J.R., Ring muhly-A grass that grows in circles, CRSQ 35(4):193–199, 1999. Return to Text.
- Bar-Yosef, O. and Kislev, M.E., Earliest domesticated barley in the Jordan Valley, National Geographic Research 2(2):257, 1986. Return to Text.



Readers’ comments
Comments are automatically closed 14 days after publication.