E. coli’s electric motor: a marvel of design

Did you know that the ‘simple’ bacterium, E. coli, swims through our gut using sophisticated, nanoscale, electric motors? Each of these motors spins a whip-like flagellum at the super-high speed of up to 22,800 revolutions per minute (some bacteria reach 102,000 rpm!1). The spinning flagellum acts as a propeller. E. coli’s motor is about 45 nm (nanometres, billionths of a metre) wide—2,000 could be lined up across the width of a human hair! Evolutionists have called it a “remarkable nanomachine”,2 “a sophisticated rotary motor”,3 and “an example of elegance in molecular engineering.”4
Motor parts
The motor is composed of parts that perform similar functions to the ones found in human-designed motors. This includes gears, a rotor, axles, a driveshaft, bushings, a universal joint, and adaptor rings (figure 1). And, just like our motors, the bacterial flagellum is powered by electricity. The bacterial cell membrane functions as a highly efficient capacitor that keeps positive and negative charges apart. This electrical difference is created by ‘proton pumps’ that take hydrogen ions (protons) and remove them from the cell. These positively charged particles then flow back into the cell, acting like an electric current. (It differs from a household electric current, which is a flow of negatively charged electrons.) As these protons flow through the flagellar motor, they cause it to spin. At the base of the motor is a central gear surrounded by up to 11 powered gears (in other flagellated bacterial species, as many as 18). The ‘current’ flowing through the powered gears causes them to rotate, driving the central gear. The powered gears rotate around axles that are anchored to the cell wall.

The rotor (figure 2) connects the central gear to the driveshaft. It is made of an inner ring, an outer ring, an adaptor ring, and a socket that houses the driveshaft.
The driveshaft passes outside the cell through a ring of proteins that serve as bushings. The outer surface of the driveshaft and inner surface of the bushings are each very smooth, and separated by a thin layer of fluid. These “optimally designed”5 bushings allow the driveshaft to rotate with very little friction, making the motor almost 100% efficient.
The hook-shaped universal joint has about 120 moving parts, each a highly specialized protein, that smoothly expand and contract as it rotates. This design makes it very resistant to twisting but very flexible to bend; two important requirements for an efficient universal joint.
Gear mechanisms
The motor has two gear-shifting mechanisms; one to change the direction of rotation and one to adjust power output. The motor switches between forward and reverse gear in less than a millisecond by changing the diameter of the top of the central gear (figure 3).

The second gear mechanism performs the same function as gears on a bicycle. If the cell swims into thicker fluid it becomes harder to rotate the propellers. The motor detects this using torque sensors, and automatically engages more powered gears with the central gear, increasing the rotational force produced by the motor.6 The opposite occurs if the cell swims into thinner fluid. This sophisticated gear mechanism makes the whole system extremely energy efficient since it doesn’t use any more power than needed.
E. coli’s navigation system
This amazing motor would be largely useless without a way to steer the cell. The cell navigates its environment using a system that tracks chemical gradients with “exquisite precision”.1 The navigation system is able to turn the cell by switching the rotation direction of the motor in any of its 5–10 flagella.
This system’s sophisticated signalling device2 has a beautiful, highly organized, hexagonal architecture (figure 4). It computes thousands of binary input signals from a vast array of sensors at the front of the device that detect different chemicals. The circuitry of the device includes a short-term memory feedback loop, allowing it to compare chemical concentrations over time. Different sensor types communicate with one another to amplify signals up to 50-fold. These features give the navigation system high sensitivity, a huge dynamic range, and an impressive amount of signal amplification.3 Yet, E. coli’s navigation system is one of the simplest in bacteria; most are much more complex.4

References and notes
- Gao, Q. and 2 others, Conformational shifts in a chemoreceptor helical hairpin control kinase signaling in Escherichia coli, pnas.org, 17 Jul 2019.
- Hazelbauer, G. and 2 others, Bacterial chemoreceptors: high-performance signaling in networked arrays, sciencedirect.com, 14 Sep 2007.
- Liu, J. and 5 others, Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells, pnas.org, 5 Jun 2012.
- Porter, S. and 2 others, Rhodobacter sphaeroides: complexity in chemotactic signalling: cell.com, 1 Jun 2008.
Motor construction
The complexity of constructing E. coli’s electric motor far exceeds the complexity of the motor itself. Numerous machines, motors, and proteins are required to manufacture, transport, and assemble each of the motor’s parts into their precise location at the specific time needed. Besides the complicated system that locates the ‘flagellum’ genes and makes proteins from them via transcription and translation, the cell has to:
- Produce a chemical fuel (ATP) to power many of the assembly machines
- Transport newly manufactured proteins to an export machine and protect them on the journey
- Cut a hole in the cell wall for the driveshaft to pass through, without bursting the cell
- Form a temporary scaffold to assist the assembly of other proteins.
And that’s just a start. Many of these tasks cannot be performed by single machines but require the collaboration of numerous machines along complicated production lines. Some of these machines also have proofreading systems which check that their tasks are performed correctly, and sometimes call on other repair machines to fix mistakes.
The proteins are manufactured with identification tags used to identify, sort, and export the proteins. The export machine controls the timing of delivery for each protein and communicates with the machines at the start of the protein production line so that a backlog is not generated. The export machine also has a built-in rotary motor that unfolds the proteins. Other machines at the assembly site refold the proteins into place.
Motor construction has three stages. In stage one, a genetic program (figure 5) takes numerous input signals from sensors which detect the cell’s acidity, oxygen levels, temperature, salinity and other factors, as well as communication signals from other bacterial cells. It uses this signal stream to decide if conditions are right to make a flagellum. Next, the core structures of the motor and the hook are assembled. Then the propeller filament, the navigation system, and the powered gears are assembled. The last step of this second stage is the construction of the hook, which, to work most efficiently, must be made to 55 nm in length.7 While being made, the export machine uses a molecular ruler to measure the length of the hook.
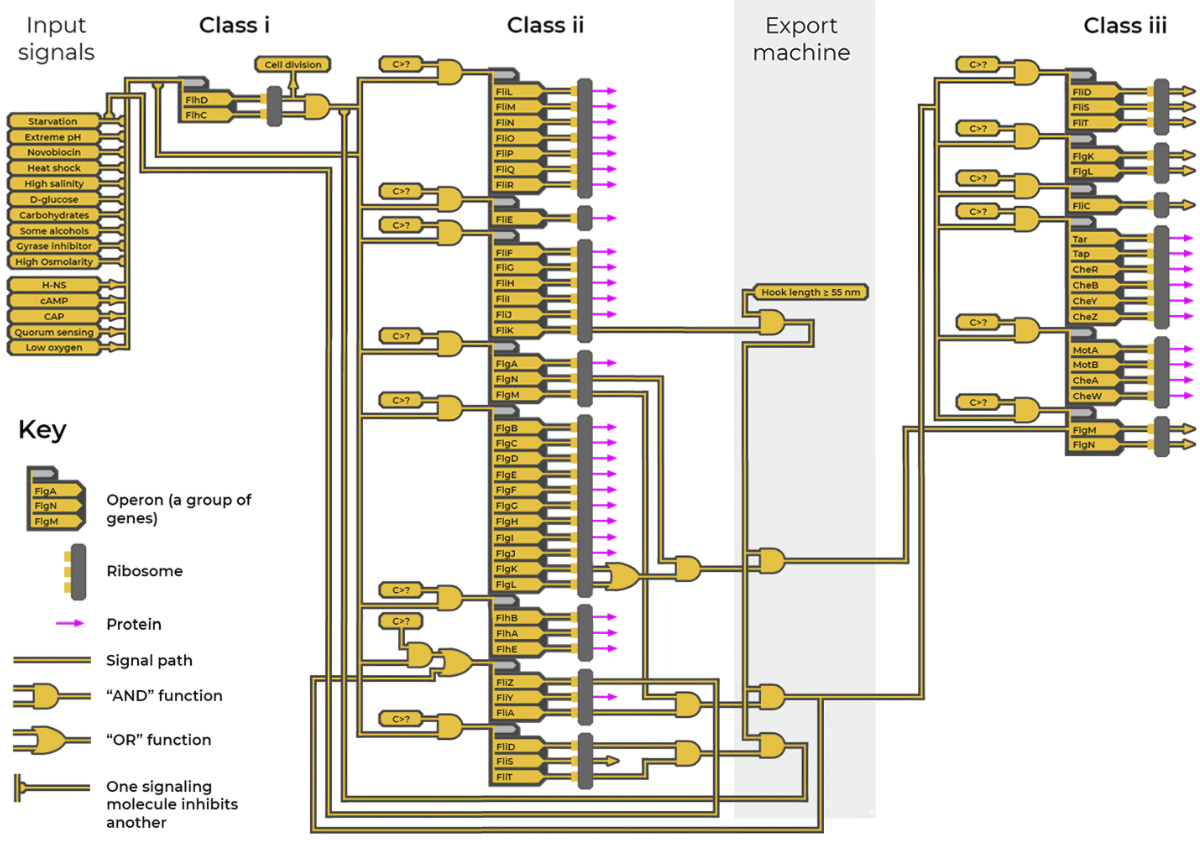
The genetic construction program

The timing and logistics of the construction process are overseen by another sophisticated program (figure 58). This genetic program functions much like a computer program with circuitry, feed-forward and feed-back loops, input and output signals, and logic gate functions.
The genetic program includes over 50 genes organized into groups (‘operons’), which are further organized into three classes. Genes contain instructions on how to make particular proteins. A protein produced from one gene can switch other genes in the program on or off, forming genetic circuitry. The production of different proteins is prioritized according to when they are required at the assembly site.
An evolutionary impossibility
Despite the clear evidence of design, many evolutionists persist in claiming that the flagellar motor evolved through random mutation and selection. Evolutionists used to claim that the flagellum evolved from an injectisome (a needle-like apparatus used to inject proteins into other cells). This is still taught in some textbooks but has now largely been rejected, because the injectisome is now thought to have evolved from the flagellum through the loss of genes. However, a recent paper9 published in April 2021 challenged both these ideas. It compared the atomic-level structure of the two and found them to be “significantly different” meaning that it would be much harder for one to evolve into the other.
A recent press release for a current study stated: “For evolutionary biologists, the flagellum … is an enduring mystery. This ‘molecular machine’ consists of many different parts that are all essential for the flagellum to work. How can gradual, Darwinian evolution build such a structure if not by inventing all necessary parts at once?”10
Conclusion
The electric motors of many bacterial species are even more complex than the one found in E. coli, with many extra parts, sometimes including a clutch11 or a brake.12 Many of the details in this article were only discovered over the last few years. No doubt, many more fascinating discoveries will be made in the years to come. Bacteria are far from ‘simple bags of chemicals’. The function, control, and construction of E. coli’s electric motor show layers upon layers of sophisticated design and engineering. This motor brings glory to its designer, our Creator, Jesus Christ (Colossians 1:16).
Figure 1:
Unless stated otherwise (see below), protein structures are sourced from the RCSB Protein Data Bank (rcsb.org). Protein graphics made using Mol* (D. Sehnal, S. Bittrich, M. Deshpande, R. Svobodová, K. Berka, V. Bazgier, S. Velankar, S.K. Burley, J. Koča, A.S. Rose (2021). Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Research. doi.org/10.1093/nar/gkab314).
PDB ID: 6SCN, Johnson, S., Fong, Y.H., Deme, J.C. et al. (2020). Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nat Microbiol, 5, 966–975 (2020). doi.org/10.1038/s41564-020-0703-3
PDB ID: 6K3I, Shibata, S., Matsunami, H., Aizawa, SI. et al. (2019). Torque transmission mechanism of the curved bacterial flagellar hook revealed by cryo-EM. Nat Struct Mol Biol, 26, 941–945. doi.org/10.1038/s41594-019-0301-3
PDB ID: 5WJY, Wang, F., Burrage, A.M., Postel, S. et al. (2017). A structural model of flagellar filament switching across multiple bacterial species. Nat Commun, 8, 960. doi.org/10.1038/s41467-017-01075-5
PDB ID: 2ZVY, Kojima, S., Imada, K., Sakuma, M., Sudo, Y., Kojima, C., Minamino, T., Homma, M. & Namba, K. (2009). Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Molecular Microbiology, 73, 710-718. doi.org/10.1111/j.1365-2958.2009.06802.x
PDB ID: 6YSL, Deme, J.C., Johnson, S., Vickery, O. et al. (2020). Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol, 5, 1553–1564. doi.org/10.1038/s41564-020-0788-8
PDB ID: 7CLR, Yamaguchi, T., Makino, F., Miyata, T. et al. (2021). Structure of the molecular bushing of the bacterial flagellar motor. Nat Commun, 12, 4469. doi.org/10.1038/s41467-021-24715-3
PDB ID: 7E80, Tan, J., Zhang, X., Wang, X., Xu, C., Chang, S., Wu, H., Wang, T., Liang, H., Gao, H., Zhou, Y., & Zhu, Y. (2021). Structural basis of assembly and torque transmission of the bacterial flagellar motor. Cell, 184(10), 2665–2679.e19. doi.org/10.1016/j.cell.2021.03.057
PDB ID: 3CX5, Solmaz, S. R., & Hunte, C. (2008). Structure of complex III with bound cytochrome c in reduced state and definition of a minimal core interface for electron transfer. The Journal of biological chemistry, 283(25), 17542–17549. doi.org/10.1074/jbc.M710126200
Image of the central gear (modified, cropped and colour change) sourced under the terms of the Creative Commons Attribution License (creativecommons.org/licenses/by/4.0/) from Brittany, L., C., Nishikino, T., Guo, W., Zhu, S., Kojima, S., Homma, M., & Liu, J. (2020). The flagellar motor of Vibrio alginolyticus undergoes major structural remodeling during rotational switching. eLife, 2020;9:e61446. doi.org/10.7554/eLife.61446
Image of the hook-filament junction (modified, cropped and colour change) sourced under the terms of the Creative Commons Attribution License from Green, A.G., Elhabashy, H., Brock, K.P. et al. (2021). Large-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences. Nat Commun, 12, 1396. doi.org/10.1038/s41467-021-21636-z
Figure 2:
Rotor protein complex structure sourced from the RCSB Protein Data Bank (rcsb.org). Protein graphic made using Mol* (D. Sehnal, S. Bittrich, M. Deshpande, R. Svobodová, K. Berka, V. Bazgier, S. Velankar, S.K. Burley, J. Koča, A.S. Rose (2021) Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Research. doi.org/10.1093/nar/gkab314).
PDB ID: 6SCN, Johnson, S., Fong, Y.H., Deme, J.C. et al. (2020) Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nat Microbiol, 5, 966–975. doi.org/10.1038/s41564-020-0703-3
References and notes
- Magariyama, Y. and 6 others, Very fast flagellar rotation. Nature 371:752, 1994. Return to text.
- Chang, Y. and 8 others, Molecular Mechanism for Rotational Switching of the Bacterial Flagellar Motor. Nature Structural & Molecular Biology, nature.com, 7 Sep 2020. Return to text.
- Baker, M. and 16 others, Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor,. Nature Structural & Molecular Biology, nature.com, 8 Feb 2016. Return to text.
- Apel, D. and Surette, M.G., Bringing order to a complex molecular machine: The assembly of the bacterial flagella, BBA Biomembranes, sciencedirect.com, Sep 2008. Return to text.
- Yamaguchi, T. and 5 others, Structure of the molecular bushing of the bacterial flagellar motor, Nat. Commun., nature.com, 22 Jul 2021. Return to text.
- Baker, A. and O’Toole, G., Bacteria, rev your engines: stator dynamics regulate flagellar motility, Journal of Bacteriology, journals.asm.org, 25 May 2017. Return to text.
- Spöring I. and 13 others, Hook length of the bacterial flagellum is optimized for maximal stability of the flagellar bundle, journals.plos.org, 6 Sep 2018. Return to text.
- Fitzgerald, D. and 2 others, Comprehensive Mapping of the Escherichia coli Flagellar Regulatory Network, journals.plos.org, 2 Oct 2014. Return to text.
- Buggs, R., More obsolete Dawkinsian evidence for evolution, natureecoevocommunity.nature.com, 4 May 2021. Return to text.
- Dunning, H., Team investigating the evolution of bacterial ‘tails’ wins prestigious grant, imperial.ac.uk, 12 Apr 2021. Return to text.
- Sarfati, J., Germ with seven motors in one!, creation.com/germ-7-motors-in-1, 15 Jan 2013. Return to text.
- Pilizota, T. and 5 others, A molecular brake, not a clutch, stops the Rhodobacter sphaeroides flagellar motor, pnas.org, 14 Jul 2009. Return to text.












Readers’ comments
Comments are automatically closed 14 days after publication.