Copying enzyme design to make industrial catalysts
What does ‘directed evolution’ really mean?

Enzymes are a vital class of proteins that are catalysts. That is, they speed up chemical reactions without being consumed in the process. Without enzymes, many reactions essential for life would be far too slow for life to exist.
For example, a reaction “absolutely essential” in creating the building blocks of DNA and RNA would take 78 million years in water. But an enzyme (orotidine 5′-monophosphate decarboxylase) makes this reaction go 1018 times faster,1 so it takes less than 3 milliseconds in the cell.2 Another enzyme (inositol phosphatase) speeds up a reaction vital in cell signalling and regulation by 1021 times. With the enzyme, the reaction takes 10 milliseconds; without the enzyme, it would take a trillion years. This is almost a hundred times the supposed evolutionary age of the universe (about 15 billion years)!3
There are also many essential enzymes for DNA repair, without which DNA would disintegrate rapidly. And the DNA would be useless unless its information could be decoded. This means that this very long and thin coiled molecule must be untangled constantly with topoisomerase enzymes. Then reading this information requires a double-sieve enzyme and many others vital for decoding and splicing the information in DNA.
This requires exquisite fine tuning of the amino acids so that the enzymes have the required shape. Indeed, the fine-tuning is at the atomic level, so the right atoms of the enzymes interact with the right atoms of the substrate. See for example the animation of the enzyme aconitase below.
Industrial catalysts
Catalysts are very important for industry as well. They are used for production of medicines and fuels, and to reduce pollution. Indeed, probably about 90% of all chemical products need catalysts at some point in their manufacture. However, industrial catalysts mainly use rare or precious metals. E.g. catalytic converters use platinum, rhodium, or palladium, which are similar in value to gold.
However, enzymes use common or earth-abundant metals (EAMs) such as magnesium, iron, and copper. If industry could make catalysts with EAMs, it would drastically lower cost and the environmental footprint. So scientists have now used enzymes from living creatures as the starting point to make industrial catalysts.4 This is one example of a rapidly expanding field called biomimetics: copying designs from nature.
Directed evolution?

Sometimes these enzymes need to be tweaked, and the process is something they call by the oxymoronic term “directed evolution”. However, this is really directed selection. That is:
- Start with enzymes from living organisms—not from random proteins.
- Mutate the genes that code for these enzymes.
- Insert genes into bacteria
- Check the resulting enzyme for desired activity
- Select the best, discard the rest.
- Go to Step 2.
This is very important chemistry, for which Dr Frances Arnold of Caltech deservedly won a half-share of the 2018 Nobel Prize for Chemistry. But is it really a triumph for evolution over creation?
Not at all! Certainly, evolution and creation are diametrically opposed ideas—did life have a Maker or not? However, many observations actually fit well in both models, so can’t be used as evidence for one over the other. This is called the Fallacy of Overlapping Predictions. So we shouldn’t be surprised that we have interviewed two creationists who have worked in this field, for example Dr Dudley Eirich and Dr Matti Leisola. Even CMI marine biologist and geneticist Dr Robert Carter used directed selection to discover new and useful protein variants as part of his doctoral work.5
Directed selection provides no support for evolution
In particular, evolutionists often ignore the fact that natural and artificial selection are part of the creation model, as creationists both before and after Darwin understood. Indeed, natural selection existed before the Fall! This has nothing to do with evolution from prokaryotes to professors, for several reasons (compare Genetic algorithms—do they show that evolution works?):
Natural selection is not so much survival of the fittest but reproduction of the fittest. Many enzymes are needed for a reproducing cell to exist. Therefore natural selection cannot explain the origin of these enzymes. So we are left only with undirected chemistry, which goes in the wrong direction:
- Small molecules would not join up to big molecules. Rather, big molecules break down into small ones.
- Even if small molecules could join together, they would do so randomly,not intospecific sequences.
- The ‘building blocks’ would be a mixture of left- and right-handed instead of100% one-handed.
- Building blocks would break down over time.
- Directed selection has a goal, e.g. increased catalytic activity or stability. Natural selection is ‘blind’.
- Directed selection is perfect. Only the best catalysts are taken to the next level, while others are discarded, i.e. extreme truncation selection. Real-world natural selection is nothing like that. Darwin himself talked about slight variations with only slight selective advantages. And this needs many generations to work, which is why long ages are necessary, although not sufficient, for evolution.
- In these experiments, the artificial ‘reproductive’ rates are enormous. Nothing like this exists in real-world biology.
Mutation rates are also extremely high in these lab trials. They can afford to be, because they are dealing with only one gene out of thousands (E. coli has 4,401 genes). Also, the chemists ensure that something survives. But if the whole bacterial genome were subjected to such a high mutation rate, it would fall apart. Even much lower mutation rates of whole genomes can cause extinctions, such as a strain of the H1N1 influenza virus.
Computer modelling shows that even truncation selection would not prevent overall degradation if the whole organism were subject to high mutation rates. See the paper Mutations: evolution’s engine becomes evolution’s end! under Truncation selection.
Related to this is the claim that random DNA sequences can produce novel functions. However, Dr Carter analyzed the claims very carefully and pointed out:
By introducing random RNAs into the cell, Neme et al. inadvertently changed the genomic regulation patterns of already existing genes. No new functions were added. No evolution has taken place. While the experiment was ingenious, the conclusions they derived from it were unwarranted. Venema [Dr Denis Venema, of theistic evolutionary propaganda group BioLogos] was premature in his praise.
- Finally, in real life, mutations are not random.
Work to be done
One problem with enzymes is that they are quite easily denatured. That is, their fine-tuned spatial arrangement can be disrupted, so they no longer function. This is a major reason why cooking kills bacteria—their enzymes are all denatured, so the cells can’t work.
However, many industrial chemical processes have conditions that are too harsh for most enzymes to survive. For example, high temperatures and pressures, highly acid or alkaline conditions, and the presence of organic solvents. Thus, with more directed selection, they hope to improve enzyme stability.
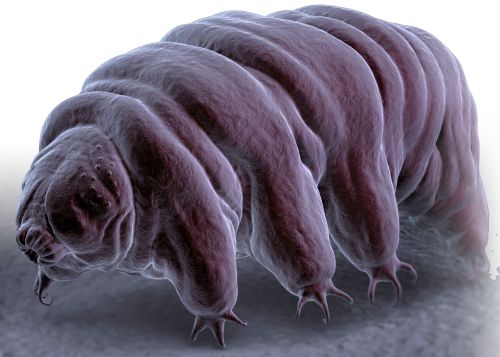
As a counter-suggestion, the living world has some creatures that can withstand extremes, so must have some way to protect their enzymes. Foremost are tardigrades. They can survive in boiling water, frigid liquid helium, six times the pressure of the deepest part of our oceans, and radiation doses 250 times the lethal dose for humans. And there are other extremophiles.
Another idea: instead of modifying the enzymes to fit the process, do the reverse. That is, improve the process to make it more amenable to work with biology. For example, the production of the strongest man-made fibre, Kevlar®, requires harsh conditions, including boiling sulfuric acid. But spider silk is even stronger, and obviously made at ordinary temperatures, and uses only mildly acidic conditions that are not harmful to spiders. Clearly, tough materials can be made without harsh conditions. All we have to do is learn more from the Creator.
Conclusion
This work highlights the brilliant design of our enzymes. And it provides a huge head start to chemists trying to develop industrial catalysts. So far, it is beyond our capacity to design an enzyme from scratch. Just predicting the 3D structure of a folded protein from its amino acid sequence “is a longstanding challenge in computational biology.”6
Also, directed selection is good operational science. It provides not the slightest evidence that random chemicals could have formed the first enzymes without intelligence. But the term ‘directed evolution’ is hardly the first time where an alleged proof of ‘evolution’ is really a proof of natural selection.
References and notes
- Miller, B.G. and 4 others, Anatomy of a proficient enzyme: The structure of orotidine 5′-monophosphate decarboxylase in the presence and absence of a potential transition state analog, PNAS 97(5):2011–2016, 2000. Return to text.
- Lead researcher Dr Richard Wolfenden; cited in: Without enzyme catalyst, slowest known biological reaction takes 1 trillion years, sciencedaily.com, 6 May 2003. Return to text.
- Lad, C., Williams, N.H., and Wolfenden, R., The rate of hydrolysis of phosphomonoester dianions and the exceptional catalytic proficiencies of protein and inositol phosphatases, PNAS 100(10):5607–5610, 2003. Return to text.
- Bullock, R.M. and 17 others, Using nature’s blueprint to expand catalysis with earth-abundant metals, Science 369(6505):786, 14 Aug 2020. Return to text.
- Carter, R.W., Schmale, M.C., and Gibbs, P.D.L., Cloning of anthozoan fluorescent protein genes, Comp Biochem Physiol C Toxicol Pharmacol 138(3):259–70, 2004; doi:10.1016/j.cca.2004.07.002. Return to text.
- Yang, J and 5 others, Improved protein structure prediction using predicted interresidue orientations, PNAS 117(3) 1496–1503, 21 Jan 2020. Return to text.




Readers’ comments
Comments are automatically closed 14 days after publication.